An algebraic language for RNA pseudoknots comparison
- PMID: 30999864
- PMCID: PMC6471698
- DOI: 10.1186/s12859-019-2689-5
An algebraic language for RNA pseudoknots comparison
Abstract
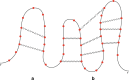
Background: RNA secondary structure comparison is a fundamental task for several studies, among which are RNA structure prediction and evolution. The comparison can currently be done efficiently only for pseudoknot-free structures due to their inherent tree representation.
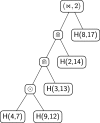
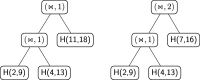
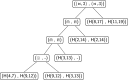
Results: In this work, we introduce an algebraic language to represent RNA secondary structures with arbitrary pseudoknots. Each structure is associated with a unique algebraic RNA tree that is derived from a tree grammar having concatenation, nesting and crossing as operators. From an algebraic RNA tree, an abstraction is defined in which the primary structure is neglected. The resulting structural RNA tree allows us to define a new measure of similarity calculated exploiting classical tree alignment.
Conclusions: The tree grammar with its operators permit to uniquely represent any RNA secondary structure as a tree. Structural RNA trees allow us to perform comparison of RNA secondary structures with arbitrary pseudoknots without taking into account the primary structure.
Keywords: ASPRA distance; Algebraic RNA tree; Structural RNA tree; Tree alignment; Tree grammar.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

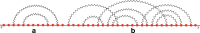



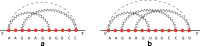


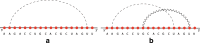






References
-
- Waterman MS. Studies on Foundations and Combinatorics, Advances in Mathematics Supplementary Studies, vol. 1. New York: Academic Press, Inc.; 1978. Secondary Structure of Single-Stranded Nucleic Acids.
-
- Waterman MS, Smith TF. RNA secondary structure: a complete mathematical analysis. Math Biosci. 1978;42(3-4):257–66.
-
- Dam ET, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31(47):11665–76. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

