Newcastle disease virus selectively infects dividing cells and promotes viral proliferation
- PMID: 30999941
- PMCID: PMC6472075
- DOI: 10.1186/s13567-019-0644-0
Newcastle disease virus selectively infects dividing cells and promotes viral proliferation
Abstract
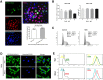
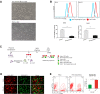
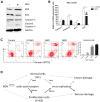
Newcastle disease virus (NDV) can select cells to infect, but the mechanism of its cell selectivity has not been comprehensively investigated. Here, we use HeLa cells to establish that NDV can selectively infect cells at the single-cell level. We labeled proliferating cells with 5'-bromo-2-deoxyuridine (BrdU) and examined the colocalization of BrdU with NDV in cells to clarify the relationships between NDV infection and cell proliferation. Receptors at the plasma membrane mediate NDV entry into host cells. We labeled sialic acid receptor isoforms, compared their densities between different cell types and measured the sialic acid receptor densities in different cell phases. Our results suggest that NDV displays host tropism to HeLa cells compared to BHK cells and that the differences in the receptor isoform expression patterns between cell types contribute to the selection of HeLa by NDV. At the single-cell level, the dynamics of receptor expression changes during different cell phases contributing to the selection of cells in S/G2 phase for NDV infection. Furthermore, cell proliferation benefits viral replication, and enhanced virus replication leads to increased damage to cells. The elucidation of the mechanisms underlying host cell selection by NDV may help in the screening and characterizing of additional candidate oncolytic virus strains.
Figures



References
-
- Ravindra PV, Tiwari AK, Sharma B, Chauhan RS. Newcastle disease virus as an oncolytic agent. Indian J Med Res. 2009;130:507–513. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

