Overlapping Roles in Chromosome Segregation for Heterochromatin Protein 1 (Swi6) and DDK in Schizosaccharomyces pombe
- PMID: 31000521
- PMCID: PMC6553818
- DOI: 10.1534/genetics.119.302125
Overlapping Roles in Chromosome Segregation for Heterochromatin Protein 1 (Swi6) and DDK in Schizosaccharomyces pombe
Abstract
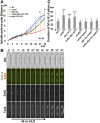
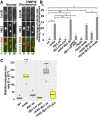
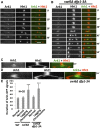
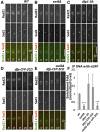
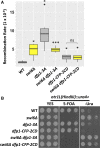
Fission yeast Swi6 is a human HP1 homolog that plays important roles in multiple cellular processes. In addition to its role in maintaining heterochromatin silencing, Swi6 is required for cohesin enrichment at the pericentromere. Loss of Swi6 leads to abnormal mitosis, including defects in the establishment of bioriented sister kinetochores and microtubule attachment. Swi6 interacts with Dfp1, a regulatory subunit of DBF4-dependent kinase (DDK), and failure to recruit Dfp1 to the pericentromere results in late DNA replication. Using the dfp1-3A mutant allele, which specifically disrupts Swi6-Dfp1 association, we investigated how interaction between Swi6 and Dfp1 affects chromosome dynamics. We find that disrupting the interaction between Swi6 and Dfp1 delays mitotic progression in a spindle assembly checkpoint-dependent manner. Artificially tethering Dfp1 back to the pericentromere is sufficient to restore normal spindle length and rescue segregation defects in swi6-deleted cells. However, Swi6 is necessary for centromeric localization of Rad21-GFP independent of DDK. Our data indicate that DDK contributes to mitotic chromosome segregation in pathways that partly overlap with, but can be separated from both, Swi6 and the other HP1 homolog, Chp2.
Keywords: Cdc7/Hsk1; Chp1 and SAC; Cnp1; fission yeast; heterochromatin; kinetochore; minichromosome; mitosis.
Copyright © 2019 by the Genetics Society of America.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

