Post-translational regulation of ubiquitin signaling
- PMID: 31000580
- PMCID: PMC6548142
- DOI: 10.1083/jcb.201902074
Post-translational regulation of ubiquitin signaling
Abstract
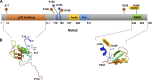
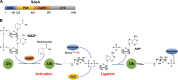
Ubiquitination regulates many essential cellular processes in eukaryotes. This post-translational modification (PTM) is typically achieved by E1, E2, and E3 enzymes that sequentially catalyze activation, conjugation, and ligation reactions, respectively, leading to covalent attachment of ubiquitin, usually to lysine residues of substrate proteins. Ubiquitin can also be successively linked to one of the seven lysine residues on ubiquitin to form distinctive forms of polyubiquitin chains, which, depending upon the lysine used and the length of the chains, dictate the fate of substrate proteins. Recent discoveries revealed that this ubiquitin code is further expanded by PTMs such as phosphorylation, acetylation, deamidation, and ADP-ribosylation, on ubiquitin, components of the ubiquitination machinery, or both. These PTMs provide additional regulatory nodes to integrate development or insulting signals with cellular homeostasis. Understanding the precise roles of these PTMs in the regulation of ubiquitin signaling will provide new insights into the mechanisms and treatment of various human diseases linked to ubiquitination, including neurodegenerative diseases, cancer, infection, and immune disorders.
© 2019 Song and Luo.
Figures




References
-
- Blomstrom D.C., Fahey D., Kutny R., Korant B.D., and Knight E. Jr. 1986. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J. Biol. Chem. 261:8811–8816. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

