GSK2801, a BAZ2/BRD9 Bromodomain Inhibitor, Synergizes with BET Inhibitors to Induce Apoptosis in Triple-Negative Breast Cancer
- PMID: 31000582
- PMCID: PMC6610760
- DOI: 10.1158/1541-7786.MCR-18-1121
GSK2801, a BAZ2/BRD9 Bromodomain Inhibitor, Synergizes with BET Inhibitors to Induce Apoptosis in Triple-Negative Breast Cancer
Abstract
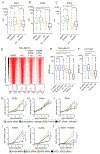
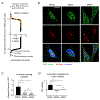
Screening of an inhibitor library targeting kinases and epigenetic regulators identified several molecules having antiproliferative synergy with extraterminal domain (BET) bromodomain (BD) inhibitors (JQ1, OTX015) in triple-negative breast cancer (TNBC). GSK2801, an inhibitor of BAZ2A/B BDs, of the imitation switch chromatin remodeling complexes, and BRD9, of the SWI/SNF complex, demonstrated synergy independent of BRD4 control of P-TEFb-mediated pause-release of RNA polymerase II. GSK2801 or RNAi knockdown of BAZ2A/B with JQ1 selectively displaced BRD2 at promoters/enhancers of ETS-regulated genes. Additional displacement of BRD2 from rDNA in the nucleolus coincided with decreased 45S rRNA, revealing a function of BRD2 in regulating RNA polymerase I transcription. In 2D cultures, enhanced displacement of BRD2 from chromatin by combination drug treatment induced senescence. In spheroid cultures, combination treatment induced cleaved caspase-3 and cleaved PARP characteristic of apoptosis in tumor cells. Thus, GSK2801 blocks BRD2-driven transcription in combination with BET inhibitor and induces apoptosis of TNBC. IMPLICATIONS: Synergistic inhibition of BDs encoded in BAZ2A/B, BRD9, and BET proteins induces apoptosis of TNBC by a combinatorial suppression of ribosomal DNA transcription and ETS-regulated genes.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

