Self-organization of Plk4 regulates symmetry breaking in centriole duplication
- PMID: 31000710
- PMCID: PMC6472344
- DOI: 10.1038/s41467-019-09847-x
Self-organization of Plk4 regulates symmetry breaking in centriole duplication
Abstract
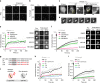
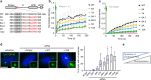
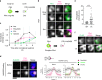
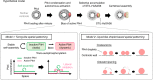
During centriole duplication, a single daughter centriole is formed next to the mother centriole. The molecular mechanism that determines a single duplication site remains a long-standing question. Here, we show that intrinsic self-organization of Plk4 is implicated in symmetry breaking in the process of centriole duplication. We demonstrate that Plk4 has an ability to phase-separate into condensates via an intrinsically disordered linker and that the condensation properties of Plk4 are regulated by autophosphorylation. Consistently, the dissociation dynamics of centriolar Plk4 are controlled by autophosphorylation. We further found that autophosphorylated Plk4 is already distributed as a single focus around the mother centriole before the initiation of procentriole formation, and is subsequently targeted for STIL-HsSAS6 loading. Perturbation of Plk4 self-organization affects the asymmetry of centriolar Plk4 distribution and proper centriole duplication. Overall, we propose that the spatial pattern formation of Plk4 is a determinant of a single duplication site per mother centriole.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Autophosphorylation-induced self-assembly and STIL-dependent reinforcement underlie Plk4's ring-to-dot localization conversion around a human centriole.Cell Cycle. 2020 Dec;19(24):3419-3436. doi: 10.1080/15384101.2020.1843772. Epub 2020 Dec 15. Cell Cycle. 2020. PMID: 33323015 Free PMC article.
-
Bimodal Binding of STIL to Plk4 Controls Proper Centriole Copy Number.Cell Rep. 2018 Jun 12;23(11):3160-3169.e4. doi: 10.1016/j.celrep.2018.05.030. Cell Rep. 2018. PMID: 29898389
-
Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole.Nat Commun. 2014 Oct 24;5:5267. doi: 10.1038/ncomms6267. Nat Commun. 2014. PMID: 25342035 Free PMC article.
-
Emerging insights into symmetry breaking in centriole duplication: updated view on centriole duplication theory.Curr Opin Struct Biol. 2021 Feb;66:8-14. doi: 10.1016/j.sbi.2020.08.005. Epub 2020 Sep 18. Curr Opin Struct Biol. 2021. PMID: 32956908 Review.
-
The PLK4-STIL-SAS-6 module at the core of centriole duplication.Biochem Soc Trans. 2016 Oct 15;44(5):1253-1263. doi: 10.1042/BST20160116. Biochem Soc Trans. 2016. PMID: 27911707 Free PMC article. Review.
Cited by
-
Protein phase separation: new insights into cell division.Acta Biochim Biophys Sin (Shanghai). 2023 May 30;55(7):1042-1051. doi: 10.3724/abbs.2023093. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37249333 Free PMC article.
-
Developmental and tissue-specific roles of mammalian centrosomes.FEBS J. 2025 Feb;292(4):709-726. doi: 10.1111/febs.17212. Epub 2024 Jun 27. FEBS J. 2025. PMID: 38935637 Free PMC article. Review.
-
Centrosome linker diversity and its function in centrosome clustering and mitotic spindle formation.EMBO J. 2023 Sep 4;42(17):e109738. doi: 10.15252/embj.2021109738. Epub 2023 Jul 4. EMBO J. 2023. PMID: 37401899 Free PMC article.
-
Amplified centrosomes-more than just a threat.EMBO Rep. 2024 Oct;25(10):4153-4167. doi: 10.1038/s44319-024-00260-0. Epub 2024 Sep 16. EMBO Rep. 2024. PMID: 39285247 Free PMC article. Review.
-
Duplication and Segregation of Centrosomes during Cell Division.Cells. 2022 Aug 7;11(15):2445. doi: 10.3390/cells11152445. Cells. 2022. PMID: 35954289 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

