The response of Sphingopyxis granuli strain TFA to the hostile anoxic condition
- PMID: 31000749
- PMCID: PMC6472365
- DOI: 10.1038/s41598-019-42768-9
The response of Sphingopyxis granuli strain TFA to the hostile anoxic condition
Abstract
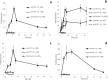
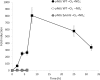
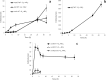
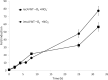
Sphingomonads comprises a group of interesting aerobic bacteria because of their ubiquity and metabolic capability of degrading many recalcitrant contaminants. The tetralin-degrader Sphingopyxis granuli strain TFA has been recently reported as able to anaerobically grow using nitrate as the alternative electron acceptor and so far is the only bacterium with this ability within the sphingomonads group. To understand how strain TFA thrives under anoxic conditions, a differential transcriptomic analysis while growing under aerobic or anoxic conditions was performed. This analysis has been validated and complemented with transcription kinetics of representative genes of different functional categories. Results show an extensive change of the expression pattern of this strain in the different conditions. Consistently, the most induced operon in anoxia codes for proteases, presumably required for extensive changes in the protein profile. Besides genes that respond to lack of oxygen in other bacteria, there are a number of genes that respond to stress or to damage of macromolecules, including genes of the SOS DNA-damage response, which suggest that anoxic conditions represent a hostile environment for this bacterium. Interestingly, growth under anoxic conditions also resulted in repression of all flagellar and type IV pilin genes, which suggested that this strain shaves its appendages off while growing in anaerobiosis.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Understanding the metabolism of the tetralin degrader Sphingopyxis granuli strain TFA through genome-scale metabolic modelling.Sci Rep. 2020 May 26;10(1):8651. doi: 10.1038/s41598-020-65258-9. Sci Rep. 2020. PMID: 32457330 Free PMC article.
-
Identification of two fnr genes and characterisation of their role in the anaerobic switch in Sphingopyxis granuli strain TFA.Sci Rep. 2020 Dec 3;10(1):21019. doi: 10.1038/s41598-020-77927-w. Sci Rep. 2020. PMID: 33273546 Free PMC article.
-
SuhB, a small non-coding RNA involved in catabolite repression of tetralin degradation genes in Sphingopyxis granuli strain TFA.Environ Microbiol. 2018 Oct;20(10):3671-3683. doi: 10.1111/1462-2920.14360. Epub 2018 Sep 14. Environ Microbiol. 2018. PMID: 30033661
-
Biodegradation of Tetralin: Genomics, Gene Function and Regulation.Genes (Basel). 2019 May 6;10(5):339. doi: 10.3390/genes10050339. Genes (Basel). 2019. PMID: 31064110 Free PMC article. Review.
-
The genus Sphingopyxis: Systematics, ecology, and bioremediation potential - A review.J Environ Manage. 2021 Feb 15;280:111744. doi: 10.1016/j.jenvman.2020.111744. Epub 2020 Dec 3. J Environ Manage. 2021. PMID: 33280938 Review.
Cited by
-
Two paralogous EcfG σ factors hierarchically orchestrate the activation of the General Stress Response in Sphingopyxis granuli TFA.Sci Rep. 2020 Mar 20;10(1):5177. doi: 10.1038/s41598-020-62101-z. Sci Rep. 2020. PMID: 32198475 Free PMC article.
-
An improved genome editing system for Sphingomonadaceae.Access Microbiol. 2024 May 13;6(5):000755.v3. doi: 10.1099/acmi.0.000755.v3. eCollection 2024. Access Microbiol. 2024. PMID: 38868378 Free PMC article.
-
A high-efficiency scar-free genome-editing toolkit for Acinetobacter baumannii.J Antimicrob Chemother. 2022 Nov 28;77(12):3390-3398. doi: 10.1093/jac/dkac328. J Antimicrob Chemother. 2022. PMID: 36216579 Free PMC article.
-
The functional differences between paralogous regulators define the control of the general stress response in Sphingopyxis granuli TFA.Environ Microbiol. 2022 Apr;24(4):1918-1931. doi: 10.1111/1462-2920.15907. Epub 2022 Jan 27. Environ Microbiol. 2022. PMID: 35049124 Free PMC article.
-
Genome features of a novel hydrocarbonoclastic Chryseobacterium oranimense strain and its comparison to bacterial oil-degraders and to other C. oranimense strains.DNA Res. 2023 Dec 1;30(6):dsad025. doi: 10.1093/dnares/dsad025. DNA Res. 2023. PMID: 37952165 Free PMC article.
References
-
- Takeuchi M, Hamana K, Hiraishi A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol. 2001;51:1405–1417. doi: 10.1099/00207713-51-4-1405. - DOI - PubMed

