Discovery of Shiga Toxin-Producing Escherichia coli (STEC)-Specific Bacteriophages From Non-fecal Composts Using Genomic Characterization
- PMID: 31001216
- PMCID: PMC6454146
- DOI: 10.3389/fmicb.2019.00627
Discovery of Shiga Toxin-Producing Escherichia coli (STEC)-Specific Bacteriophages From Non-fecal Composts Using Genomic Characterization
Abstract
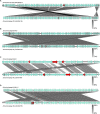
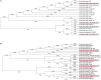
Composting is a complex biodegradable process that converts organic materials into nutrients to facilitate crop yields, and, if well managed, can render bactericidal effects. Majority of research focused on detection of enteric pathogens, such as Shiga toxin-producing Escherichia coli (STEC) in fecal composts. Recently, attention has been emphasized on bacteriophages, such as STEC-specific bacteriophages, associated with STEC from the fecal-contaminated environment because they are able to sustain adverse environmental condition during composting process. However, little is known regarding the isolation of STEC-specific bacteriophages in non-fecal composts. Thus, the objectives were to isolate and genomically characterize STEC-specific bacteriophages, and to evaluate its association with STEC in non-fecal composts. For bacteriophage isolation, the samples were enriched with non-pathogenic E. coli (3 strains) and STEC (14 strains), respectively. After purification, host range, plaque size, and phage morphology were examined. Furthermore, bacteriophage genomes were subjected to whole-genome sequencing using Illumina MiSeq and genomic analyses. Isolation of top six non-O157 and O157 STEC utilizing culture methods combined with PCR-based confirmation was also conducted. The results showed that various STEC-specific bacteriophages, including vB_EcoM-Ro111lw, vB_EcoM-Ro121lw, vB_EcoS-Ro145lw, and vB_EcoM-Ro157lw, with different but complementary host ranges were isolated. Genomic analysis showed the genome sizes varied from 42kb to 149kb, and most bacteriophages were unclassified at the genus level, except vB_EcoM-Ro111lw as FelixO1-like viruses. Prokka predicted less than 25% of the ORFs coded for known functions, including those essential for DNA replication, bacteriophage structure, and host cell lysis. Moreover, none of the bacteriophages harbored lysogenic genes or virulence genes, such as stx or eae. Additionally, the presence of these lytic bacteriophages was likely attributed to zero isolation of STEC and could also contribute to additional antimicrobial effects in composts, if the composting process was insufficient. Current findings indicate that various STEC-specific bacteriophages were found in the non-fecal composts. In addition, the genomic characterization provides in-depth information to complement the deficiency of biological features regarding lytic cycle of the new bacteriophages. Most importantly, these bacteriophages have great potential to control various serogroups of STEC.
Keywords: STEC-specific bacteriophages; Shiga toxin-producing E. coli (STEC); complementary host range; non-fecal composts; whole genome sequencing.
Figures


Similar articles
-
Characterization of Two New Shiga Toxin-Producing Escherichia coli O103-Infecting Phages Isolated from an Organic Farm.Microorganisms. 2021 Jul 17;9(7):1527. doi: 10.3390/microorganisms9071527. Microorganisms. 2021. PMID: 34361962 Free PMC article.
-
Characterization of a T4-like Bacteriophage vB_EcoM-Sa45lw as a Potential Biocontrol Agent for Shiga Toxin-Producing Escherichia coli O45 Contaminated on Mung Bean Seeds.Microbiol Spectr. 2022 Feb 23;10(1):e0222021. doi: 10.1128/spectrum.02220-21. Epub 2022 Feb 2. Microbiol Spectr. 2022. PMID: 35107386 Free PMC article.
-
Genomic Characterization of Two Shiga Toxin-Converting Bacteriophages Induced From Environmental Shiga Toxin-Producing Escherichia coli.Front Microbiol. 2021 Feb 25;12:587696. doi: 10.3389/fmicb.2021.587696. eCollection 2021. Front Microbiol. 2021. PMID: 33716997 Free PMC article.
-
A new Rogue-like Escherichia phage UDF157lw to control Escherichia coli O157:H7.Front Microbiol. 2024 Jan 22;14:1302032. doi: 10.3389/fmicb.2023.1302032. eCollection 2023. Front Microbiol. 2024. PMID: 38318127 Free PMC article.
-
Methods for the detection and isolation of Shiga toxin-producing Escherichia coli.Symp Ser Soc Appl Microbiol. 2000;(29):133S-143S. doi: 10.1111/j.1365-2672.2000.tb05341.x. Symp Ser Soc Appl Microbiol. 2000. PMID: 10880188 Review.
Cited by
-
A New Pipeline for Designing Phage Cocktails Based on Phage-Bacteria Infection Networks.Front Microbiol. 2021 Feb 16;12:564532. doi: 10.3389/fmicb.2021.564532. eCollection 2021. Front Microbiol. 2021. PMID: 33664712 Free PMC article.
-
Characterization of the novel temperate Escherichia coli phage phiStx2k.Arch Virol. 2023 Dec 11;169(1):5. doi: 10.1007/s00705-023-05941-0. Arch Virol. 2023. PMID: 38078984
-
Characterization of Two New Shiga Toxin-Producing Escherichia coli O103-Infecting Phages Isolated from an Organic Farm.Microorganisms. 2021 Jul 17;9(7):1527. doi: 10.3390/microorganisms9071527. Microorganisms. 2021. PMID: 34361962 Free PMC article.
-
Characterization of Non-O157 STEC Infecting Bacteriophages Isolated from Cattle Faeces in North-West South Africa.Microorganisms. 2019 Nov 26;7(12):615. doi: 10.3390/microorganisms7120615. Microorganisms. 2019. PMID: 31779135 Free PMC article.
-
Nettle manure: an unsuspected source of bacteriophages active against various phytopathogenic bacteria.Arch Virol. 2022 Apr;167(4):1099-1110. doi: 10.1007/s00705-022-05391-0. Epub 2022 Mar 12. Arch Virol. 2022. PMID: 35277777
References
-
- Amgarten D., Martins L. F., Lombardi K. C., Antunes L. P., De Souza A. P. S., Nicastro G. G., et al. (2017). Three novel pseudomonas phages isolated from composting provide insights into the evolution and diversity of tailed phages. BMC Genomics 18:346. 10.1186/s12864-12017-13729-z - DOI - PMC - PubMed
-
- Asare P. T., Jeong T. Y., Ryu S., Klumpp J., Loessner M. J., Merrill B. D., et al. (2015). Putative type 1 thymidylate synthase and dihydrofolate reductase as signature genes of a novel bastille-like group of phages in the subfamily Spounavirinae. BMC Genomics 16:582. 10.1186/s12864-015-1757-0 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

