Translocase of the Outer Mitochondrial Membrane 40 Is Required for Mitochondrial Biogenesis and Embryo Development in Arabidopsis
- PMID: 31001303
- PMCID: PMC6455079
- DOI: 10.3389/fpls.2019.00389
Translocase of the Outer Mitochondrial Membrane 40 Is Required for Mitochondrial Biogenesis and Embryo Development in Arabidopsis
Abstract
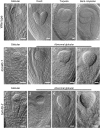
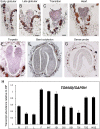
In eukaryotes, mitochondrion is an essential organelle which is surrounded by a double membrane system, including the outer membrane, intermembrane space and the inner membrane. The translocase of the outer mitochondrial membrane (TOM) complex has attracted enormous interest for its role in importing the preprotein from the cytoplasm into the mitochondrion. However, little is understood about the potential biological function of the TOM complex in Arabidopsis. The aim of the present study was to investigate how AtTOM40, a gene encoding the core subunit of the TOM complex, works in Arabidopsis. As a result, we found that lack of AtTOM40 disturbed embryo development and its pattern formation after the globular embryo stage, and finally caused albino ovules and seed abortion at the ratio of a quarter in the homozygous tom40 plants. Further investigation demonstrated that AtTOM40 is wildly expressed in different tissues, especially in cotyledons primordium during Arabidopsis embryogenesis. Moreover, we confirmed that the encoded protein AtTOM40 is localized in mitochondrion, and the observation of the ultrastructure revealed that mitochondrion biogenesis was impaired in tom40-1 embryo cells. Quantitative real-time PCR was utilized to determine the expression of genes encoding outer mitochondrial membrane proteins in the homozygous tom40-1 mutant embryos, including the genes known to be involved in import, assembly and transport of mitochondrial proteins, and the results demonstrated that most of the gene expressions were abnormal. Similarly, the expression of genes relevant to embryo development and pattern formation, such as SAM (shoot apical meristem), cotyledon, vascular primordium and hypophysis, was also affected in homozygous tom40-1 mutant embryos. Taken together, we draw the conclusion that the AtTOM40 gene is essential for the normal structure of the mitochondrion, and participates in early embryo development and pattern formation through maintaining the biogenesis of mitochondria. The findings of this study may provide new insight into the biological function of the TOM40 subunit in higher plants.
Keywords: Arabidopsis; embryo development; mitochondria biogenesis; pattern formation; translocase of the outer mitochondrial membrane 40 (TOM40).
Figures






Similar articles
-
Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway.J Biol Chem. 2004 Apr 30;279(18):18188-94. doi: 10.1074/jbc.M400050200. Epub 2004 Feb 20. J Biol Chem. 2004. PMID: 14978039
-
Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane.J Mol Biol. 2011 Jan 7;405(1):113-24. doi: 10.1016/j.jmb.2010.11.002. Epub 2010 Nov 6. J Mol Biol. 2011. PMID: 21059357
-
Arabidopsis EMB1990 Encoding a Plastid-Targeted YlmG Protein Is Required for Chloroplast Biogenesis and Embryo Development.Front Plant Sci. 2018 Feb 16;9:181. doi: 10.3389/fpls.2018.00181. eCollection 2018. Front Plant Sci. 2018. PMID: 29503657 Free PMC article.
-
How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane?J Cell Biol. 2005 Nov 7;171(3):419-23. doi: 10.1083/jcb.200507147. Epub 2005 Oct 31. J Cell Biol. 2005. PMID: 16260501 Free PMC article. Review.
-
Protein import into mitochondria of Neurospora crassa.Fungal Genet Biol. 2002 Jul;36(2):85-90. doi: 10.1016/S1087-1845(02)00018-X. Fungal Genet Biol. 2002. PMID: 12081461 Review.
Cited by
-
Genome wide association analysis for yield related traits in maize.BMC Plant Biol. 2022 Sep 21;22(1):449. doi: 10.1186/s12870-022-03812-5. BMC Plant Biol. 2022. PMID: 36127632 Free PMC article.
-
Chromosome-scale assemblies of the male and female Populus euphratica genomes reveal the molecular basis of sex determination and sexual dimorphism.Commun Biol. 2022 Nov 4;5(1):1186. doi: 10.1038/s42003-022-04145-7. Commun Biol. 2022. PMID: 36333427 Free PMC article.
-
TOMM34 serves as a candidate therapeutic target associated with immune cell infiltration in colon cancer.Front Oncol. 2023 Feb 9;13:947364. doi: 10.3389/fonc.2023.947364. eCollection 2023. Front Oncol. 2023. PMID: 36845719 Free PMC article.
-
Rapid Single-Step Affinity Purification of HA-Tagged Plant Mitochondria.Plant Physiol. 2020 Feb;182(2):692-706. doi: 10.1104/pp.19.00732. Epub 2019 Dec 9. Plant Physiol. 2020. PMID: 31818904 Free PMC article.
-
Molecular characterization, targeting and expression analysis of chloroplast and mitochondrion protein import components in Nicotiana benthamiana.Front Plant Sci. 2022 Oct 26;13:1040688. doi: 10.3389/fpls.2022.1040688. eCollection 2022. Front Plant Sci. 2022. PMID: 36388587 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

