Application of a Sensitive and Reproducible Label-Free Proteomic Approach to Explore the Proteome of Individual Meiotic-Phase Barley Anthers
- PMID: 31001307
- PMCID: PMC6454111
- DOI: 10.3389/fpls.2019.00393
Application of a Sensitive and Reproducible Label-Free Proteomic Approach to Explore the Proteome of Individual Meiotic-Phase Barley Anthers
Abstract
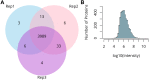
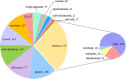
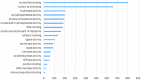
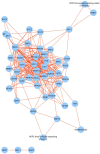
Meiosis is a highly dynamic and precisely regulated process of cell division, leading to the production of haploid gametes from one diploid parental cell. In the crop plant barley (Hordeum vulgare), male meiosis occurs in anthers, in specialized cells called meiocytes. Barley meiotic tissue is scarce and not easily accessible, making meiosis study a challenging task. We describe here a new micro-proteomics workflow that allows sensitive and reproducible genome-wide label-free proteomic analysis of individual staged barley anthers. This micro-proteomic approach detects more than 4,000 proteins from such small amounts of material as two individual anthers, covering a dynamic range of protein relative abundance levels across five orders of magnitude. We applied our micro-proteomics workflow to investigate the proteome of the developing barley anther containing pollen mother cells in the early stages of meiosis and we successfully identified 57 known and putative meiosis-related proteins. Meiotic proteins identified in our study were found to be key players of many steps and processes in early prophase such as: chromosome condensation, synapsis, DNA double-strand breaks or crossover formation. Considering the small amount of starting material, this work demonstrates an important technological advance in plant proteomics and can be applied for proteomic examination of many size-limited plant specimens. Moreover, it is the first insight into the proteome of individual barley anther at early meiosis. The proteomic data have been deposited to the ProteomeXchange with the accession number PXD010887.
Keywords: anthers; label-free micro-proteomics; mass spectrometry LC-MS/MS; meiocytes; meiosis; plants.
Figures


Similar articles
-
The proteome of developing barley anthers during meiotic prophase I.J Exp Bot. 2022 Mar 2;73(5):1464-1482. doi: 10.1093/jxb/erab494. J Exp Bot. 2022. PMID: 34758083 Free PMC article.
-
A strategy to investigate the plant meiotic proteome.Cytogenet Genome Res. 2005;109(1-3):181-9. doi: 10.1159/000082398. Cytogenet Genome Res. 2005. PMID: 15753575
-
Barley Anther and Meiocyte Transcriptome Dynamics in Meiotic Prophase I.Front Plant Sci. 2021 Jan 12;11:619404. doi: 10.3389/fpls.2020.619404. eCollection 2020. Front Plant Sci. 2021. PMID: 33510760 Free PMC article.
-
The meiotic transcriptome architecture of plants.Front Plant Sci. 2014 Jun 5;5:220. doi: 10.3389/fpls.2014.00220. eCollection 2014. Front Plant Sci. 2014. PMID: 24926296 Free PMC article. Review.
-
Premeiotic events and meiotic chromosome pairing.Symp Soc Exp Biol. 1984;38:87-121. Symp Soc Exp Biol. 1984. PMID: 6400221 Review.
Cited by
-
Shotgun Proteomics as a Powerful Tool for the Study of the Proteomes of Plants, Their Pathogens, and Plant-Pathogen Interactions.Proteomes. 2022 Jan 19;10(1):5. doi: 10.3390/proteomes10010005. Proteomes. 2022. PMID: 35225985 Free PMC article. Review.
-
The proteome of developing barley anthers during meiotic prophase I.J Exp Bot. 2022 Mar 2;73(5):1464-1482. doi: 10.1093/jxb/erab494. J Exp Bot. 2022. PMID: 34758083 Free PMC article.
-
Heat stress response mechanisms in pollen development.New Phytol. 2021 Jul;231(2):571-585. doi: 10.1111/nph.17380. Epub 2021 May 20. New Phytol. 2021. PMID: 33818773 Free PMC article. Review.
-
Ubiquitination in Plant Meiosis: Recent Advances and High Throughput Methods.Front Plant Sci. 2021 Apr 7;12:667314. doi: 10.3389/fpls.2021.667314. eCollection 2021. Front Plant Sci. 2021. PMID: 33897750 Free PMC article. Review.
-
SP3 Protocol for Proteomic Plant Sample Preparation Prior LC-MS/MS.Front Plant Sci. 2021 Mar 10;12:635550. doi: 10.3389/fpls.2021.635550. eCollection 2021. Front Plant Sci. 2021. PMID: 33777071 Free PMC article.
References
LinkOut - more resources
Full Text Sources

