Advanced Genetic Approaches in Discovery and Characterization of Genes Involved With Osteoporosis in Mouse and Human
- PMID: 31001327
- PMCID: PMC6455049
- DOI: 10.3389/fgene.2019.00288
Advanced Genetic Approaches in Discovery and Characterization of Genes Involved With Osteoporosis in Mouse and Human
Abstract
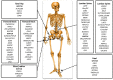
Osteoporosis is a complex condition with contributions from, and interactions between, multiple genetic loci and environmental factors. This review summarizes key advances in the application of genetic approaches for the identification of osteoporosis susceptibility genes. Genome-wide linkage analysis (GWLA) is the classical approach for identification of genes that cause monogenic diseases; however, it has shown limited success for complex diseases like osteoporosis. In contrast, genome-wide association studies (GWAS) have successfully identified over 200 osteoporosis susceptibility loci with genome-wide significance, and have provided most of the candidate genes identified to date. Phenome-wide association studies (PheWAS) apply a phenotype-to-genotype approach which can be used to complement GWAS. PheWAS is capable of characterizing the association between osteoporosis and uncommon and rare genetic variants. Another alternative approach, whole genome sequencing (WGS), will enable the discovery of uncommon and rare genetic variants in osteoporosis. Meta-analysis with increasing statistical power can offer greater confidence in gene searching through the analysis of combined results across genetic studies. Recently, new approaches to gene discovery include animal phenotype based models such as the Collaborative Cross and ENU mutagenesis. Site-directed mutagenesis and genome editing tools such as CRISPR/Cas9, TALENs and ZNFs have been used in functional analysis of candidate genes in vitro and in vivo. These resources are revolutionizing the identification of osteoporosis susceptibility genes through the use of genetically defined inbred mouse libraries, which are screened for bone phenotypes that are then correlated with known genetic variation. Identification of osteoporosis-related susceptibility genes by genetic approaches enables further characterization of gene function in animal models, with the ultimate aim being the identification of novel therapeutic targets for osteoporosis.
Keywords: GWAS; GWLA; PheWAS; WGS; collaborative cross; genome editing; osteoporosis.
Figures




References
-
- Ackert-Bicknell C. L., Karasik D., Li Q. A., Smith R. V., Hsu Y. H., Churchill G. A., et al. (2010). Mouse BMD quantitative trait loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J. Bone Miner. Res. 25 1808–1820. 10.1002/jbmr.72 - DOI - PMC - PubMed
-
- Black D. M., Cauley J. A., Wagman R., Ensrud K., Fink H. A., Hillier T. A., et al. (2018). The ability of a single BMD and fracture history assessment to predict fracture over 25 years in postmenopausal women: the study of osteoporotic fractures. J. Bone Miner. Res. 33 389–395. 10.1002/jbmr.3194 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

