Metadata Analysis Approaches for Understanding and Improving the Functional Involvement of Rumen Microbial Consortium in Digestion and Metabolism of Plant Biomass
- PMID: 31001361
- PMCID: PMC6470328
- DOI: 10.7150/jgen.32164
Metadata Analysis Approaches for Understanding and Improving the Functional Involvement of Rumen Microbial Consortium in Digestion and Metabolism of Plant Biomass
Abstract
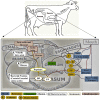
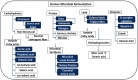
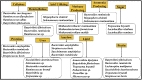
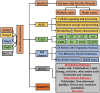
Rumen is one of the most complex gastro-intestinal system in ruminating animals. With bountiful of microorganisms supporting in breakdown and consumption of minerals and nutrients from the complex plant biomass. It is predicted that a table spoon of ruminal fluid can reside up to 150 billion microorganisms including various species of bacteria, fungi and protozoa. Several studies in the past have extensively explained about the structural and functional physiology of the rumen. Studies based on rumen and its microbiota has increased significantly in the last decade to understand and reveal applications of the rumen microbiota in food processing, pharmaceutical, biofuel and biorefining industries. Recent high-throughput meta-genomic and proteomic studies have revealed humongous information on rumen microbial diversity. In this study, we have extensively reviewed and reported present-day's progress in understanding the rumen microbial diversity. As of today, NCBI resides about 821,870 records based on rumen with approximately 889 genome sequencing studies. We have retrieved all the rumen-based records from NCBI and extensively catalogued the rumen microbial diversity and the corresponding genomic and proteomic studies respectively. Also, we have provided a brief inventory of metadata analysis software packages and reviewed the metadata analysis approaches for understanding the functional involvement of these microorganisms. Knowing and understanding the present progress on rumen microbiota and performing metadata analysis studies will significantly benefit the researchers in identifying the molecular mechanisms involved in plant biomass degradation. These studies are also necessary for developing highly efficient microorganisms and enzyme mixtures for enhancing the benefits of cattle-feedstock and biofuel industries.
Keywords: Bacteria; Fungi; Genomic; Microbiota; Proteomic; Protozoa.; Ruminal fluid; Ruminating animals.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Ishler VA, Heinrichs AJ, Varga GB. From feed to milk: understanding rumen function: Pennsylvania State University; 1996.
-
- Hungate R. The rumen microbial ecosystem. Annual Review of Ecology and Systematics. 1975;6:39–66.
-
- Hobson PN, Stewart CS. The rumen microbial ecosystem: Springer Science & Business Media; 2012.
-
- Harfoot C, Hazlewood G. Lipid metabolism in the rumen. The rumen microbial ecosystem: Springer; 1997. p. 382-426.
-
- Krehbiel C. INVITED REVIEW: Applied nutrition of ruminants: Fermentation and digestive physiology1. The Professional Animal Scientist. 2014;30:129–39.
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

