Histone Modifications Drive Aberrant Notch3 Expression/Activity and Growth in T-ALL
- PMID: 31001470
- PMCID: PMC6456714
- DOI: 10.3389/fonc.2019.00198
Histone Modifications Drive Aberrant Notch3 Expression/Activity and Growth in T-ALL
Abstract
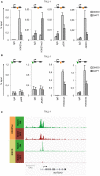
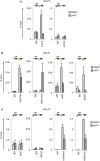
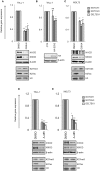
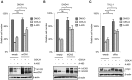
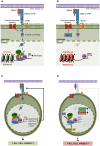
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive blood cancer caused by the deregulation of key T-cell developmental pathways, including Notch signaling. Aberrant Notch signaling in T-ALL occurs by NOTCH1 gain-of-function mutations and by NOTCH3 overexpression. Although NOTCH3 is assumed as a Notch1 target, machinery driving its transcription in T-ALL is undefined in leukemia subsets lacking Notch1 activation. Here, we found that the binding of the intracellular Notch3 domain, as well as of the activated Notch1 fragment, to the NOTCH3 gene locus led to the recruitment of the H3K27 modifiers JMJD3 and p300, and it was required to preserve transcriptional permissive/active H3K27 marks and to sustain NOTCH3 gene expression levels. Consistently, pharmacological inhibition of JMJD3 by GSKJ4 treatment or of p300 by A-485 decreased the levels of expression of NOTCH3, NOTCH1 and of the Notch target genes DELTEX1 and c-Myc and abrogated cell viability in both Notch1- and Notch3-dependent T-cell contexts. Notably, re-introduction of exogenous Notch1, Notch3 as well as c-Myc partially rescued cells from anti-growth effects induced by either treatment. Overall our findings indicate JMJD3 and p300 as general Notch1 and Notch3 signaling co-activators in T-ALL and suggest further investigation on the potential therapeutic anti-leukemic efficacy of their enzymatic inhibition in Notch/c-Myc axis-related cancers and diseases.
Keywords: A-485; GSKJ4; JMJD3; Notch signaling; T-cell lymphoblastic leukemia; epigenetics; gene regulation; p300.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

