A 192-heme electron transfer network in the hydrazine dehydrogenase complex
- PMID: 31001586
- PMCID: PMC6469936
- DOI: 10.1126/sciadv.aav4310
A 192-heme electron transfer network in the hydrazine dehydrogenase complex
Abstract
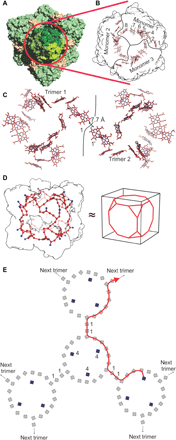
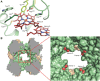
Anaerobic ammonium oxidation (anammox) is a major process in the biogeochemical nitrogen cycle in which nitrite and ammonium are converted to dinitrogen gas and water through the highly reactive intermediate hydrazine. So far, it is unknown how anammox organisms convert the toxic hydrazine into nitrogen and harvest the extremely low potential electrons (-750 mV) released in this process. We report the crystal structure and cryo electron microscopy structures of the responsible enzyme, hydrazine dehydrogenase, which is a 1.7 MDa multiprotein complex containing an extended electron transfer network of 192 heme groups spanning the entire complex. This unique molecular arrangement suggests a way in which the protein stores and releases the electrons obtained from hydrazine conversion, the final step in the globally important anammox process.
Figures

Similar articles
-
Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex.Nat Microbiol. 2021 Sep;6(9):1129-1139. doi: 10.1038/s41564-021-00934-8. Epub 2021 Jul 15. Nat Microbiol. 2021. PMID: 34267357 Free PMC article.
-
A nitric oxide-binding heterodimeric cytochrome c complex from the anammox bacterium Kuenenia stuttgartiensis binds to hydrazine synthase.J Biol Chem. 2019 Nov 8;294(45):16712-16728. doi: 10.1074/jbc.RA119.008788. Epub 2019 Sep 22. J Biol Chem. 2019. PMID: 31548310 Free PMC article.
-
Purification of the key enzyme complexes of the anammox pathway from DEMON sludge.Biopolymers. 2021 Jun;112(6):e23428. doi: 10.1002/bip.23428. Epub 2021 Apr 2. Biopolymers. 2021. PMID: 33798263
-
Anammox Biochemistry: a Tale of Heme c Proteins.Trends Biochem Sci. 2016 Dec;41(12):998-1011. doi: 10.1016/j.tibs.2016.08.015. Epub 2016 Sep 23. Trends Biochem Sci. 2016. PMID: 27669648 Review.
-
Anammox--growth physiology, cell biology, and metabolism.Adv Microb Physiol. 2012;60:211-62. doi: 10.1016/B978-0-12-398264-3.00003-6. Adv Microb Physiol. 2012. PMID: 22633060 Review.
Cited by
-
Characterization of a nitrite-reducing octaheme hydroxylamine oxidoreductase that lacks the tyrosine cross-link.J Biol Chem. 2021 Jan-Jun;296:100476. doi: 10.1016/j.jbc.2021.100476. Epub 2021 Feb 27. J Biol Chem. 2021. PMID: 33652023 Free PMC article.
-
Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex.Nat Microbiol. 2021 Sep;6(9):1129-1139. doi: 10.1038/s41564-021-00934-8. Epub 2021 Jul 15. Nat Microbiol. 2021. PMID: 34267357 Free PMC article.
-
Phylogenomic Evidence for the Origin of Obligate Anaerobic Anammox Bacteria Around the Great Oxidation Event.Mol Biol Evol. 2022 Aug 3;39(8):msac170. doi: 10.1093/molbev/msac170. Mol Biol Evol. 2022. PMID: 35920138 Free PMC article.
-
Specificity of Small c-Type Cytochromes in Anaerobic Ammonium Oxidation.ACS Omega. 2021 Aug 9;6(33):21457-21464. doi: 10.1021/acsomega.1c02275. eCollection 2021 Aug 24. ACS Omega. 2021. PMID: 34471748 Free PMC article.
-
Light-driven ammonium oxidation to dinitrogen gas by self-photosensitized biohybrid anammox systems.iScience. 2023 Apr 23;26(5):106725. doi: 10.1016/j.isci.2023.106725. eCollection 2023 May 19. iScience. 2023. PMID: 37216127 Free PMC article.
References
-
- Mulder A., van de Graaf A. A., Robertson L. A., Kuenen J. G., Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 16, 177–183 (1995).
-
- Arrigo K. R., Marine microorganisms and global nutrient cycles. Nature 437, 349–355 (2005). - PubMed
-
- van Niftrik L. A., Fuerst J. A., Damsté J. S. S., Kuenen J. G., Jetten M. S. M., Strous M., The anammoxosome: An intracytoplasmic compartment in anammox bacteria. FEMS Microbiol. Lett. 233, 7–13 (2004). - PubMed
-
- Kartal B., de Almeida N. M., Maalcke W. J., Op den Camp H. J. M., Jetten M. S. M., Keltjens J. T., How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 37, 428–461 (2013). - PubMed
-
- Dietl A., Ferousi C., Maalcke W. J., Menzel A., de Vries S., Keltjens J. T., Jetten M. S. M., Kartal B., Barends T. R. M., The inner workings of the hydrazine synthase multiprotein complex. Nature 527, 394–397 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

