Mimivirus encodes a multifunctional primase with DNA/RNA polymerase, terminal transferase and translesion synthesis activities
- PMID: 31001622
- PMCID: PMC6648351
- DOI: 10.1093/nar/gkz236
Mimivirus encodes a multifunctional primase with DNA/RNA polymerase, terminal transferase and translesion synthesis activities
Abstract
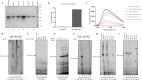
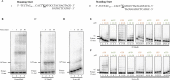
Acanthamoeba polyphaga mimivirus is an amoeba-infecting giant virus with over 1000 genes including several involved in DNA replication and repair. Here, we report the biochemical characterization of gene product 577 (gp577), a hypothetical protein (product of L537 gene) encoded by mimivirus. Sequence analysis and phylogeny suggested gp577 to be a primase-polymerase (PrimPol)-the first PrimPol to be identified in a nucleocytoplasmic large DNA virus (NCLDV). Recombinant gp577 protein purified as a homodimer and exhibited de novo RNA as well as DNA synthesis on circular and linear single-stranded DNA templates. Further, gp577 extends a DNA/RNA primer annealed to a DNA or RNA template using deoxyribonucleoties (dNTPs) or ribonucleotides (NTPs) demonstrating its DNA/RNA polymerase and reverse transcriptase activity. We also show that gp577 possesses terminal transferase activity and is capable of extending ssDNA and dsDNA with NTPs and dNTPs. Mutation of the conserved primase motif residues of gp577 resulted in the loss of primase, polymerase, reverse transcriptase and terminal transferase activities. Additionally, we show that gp577 possesses translesion synthesis (TLS) activity. Mimiviral gp577 represents the first protein from an NCLDV endowed with primase, polymerase, reverse transcriptase, terminal transferase and TLS activities.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

