Canagliflozin reduces inflammation and fibrosis biomarkers: a potential mechanism of action for beneficial effects of SGLT2 inhibitors in diabetic kidney disease
- PMID: 31001673
- PMCID: PMC6560022
- DOI: 10.1007/s00125-019-4859-4
Canagliflozin reduces inflammation and fibrosis biomarkers: a potential mechanism of action for beneficial effects of SGLT2 inhibitors in diabetic kidney disease
Abstract
Aims/hypothesis: The sodium-glucose cotransporter 2 (SGLT2) inhibitor canagliflozin slows progression of kidney function decline in type 2 diabetes. The aim of this study was to assess the effect of the SGLT2 inhibitor canagliflozin on biomarkers for progression of diabetic kidney disease (DKD).
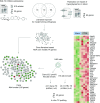
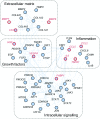
Methods: A canagliflozin mechanism of action (MoA) network model was constructed based on an in vitro transcriptomics experiment in human proximal tubular cells and molecular features linked to SGLT2 inhibitors from scientific literature. This model was mapped onto an established DKD network model that describes molecular processes associated with DKD. Overlapping areas in both networks were subsequently used to select candidate biomarkers that change with canagliflozin therapy. These biomarkers were measured in 296 stored plasma samples from a previously reported 2 year clinical trial comparing canagliflozin with glimepiride.
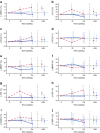
Results: Forty-four proteins present in the canagliflozin MoA molecular model overlapped with proteins in the DKD network model. These proteins were considered candidates for monitoring impact of canagliflozin on DKD pathophysiology. For ten of these proteins, scientific evidence was available suggesting that they are involved in DKD progression. Of these, compared with glimepiride, canagliflozin 300 mg/day decreased plasma levels of TNF receptor 1 (TNFR1; 9.2%; p < 0.001), IL-6 (26.6%; p = 0.010), matrix metalloproteinase 7 (MMP7; 24.9%; p = 0.011) and fibronectin 1 (FN1; 14.9%; p = 0.055) during 2 years of follow-up.
Conclusions/interpretation: The observed reduction in TNFR1, IL-6, MMP7 and FN1 suggests that canagliflozin contributes to reversing molecular processes related to inflammation, extracellular matrix turnover and fibrosis. Trial registration ClinicalTrials.gov NCT00968812.
Keywords: Biomarkers; Canagliflozin; Chronic kidney disease; Sodium–glucose cotransporter 2 inhibitors; Type 2 diabetes.
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

