Nipah Virus Sequences from Humans and Bats during Nipah Outbreak, Kerala, India, 2018
- PMID: 31002049
- PMCID: PMC6478210
- DOI: 10.3201/eid2505.181076
Nipah Virus Sequences from Humans and Bats during Nipah Outbreak, Kerala, India, 2018
Abstract
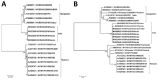
We retrieved Nipah virus (NiV) sequences from 4 human and 3 fruit bat (Pteropus medius) samples from a 2018 outbreak in Kerala, India. Phylogenetic analysis demonstrated that NiV from humans was 96.15% similar to a Bangladesh strain but 99.7%-100% similar to virus from Pteropus spp. bats, indicating bats were the source of the outbreak.
Keywords: Human; India; Kerala; Nipah virus; Pteropus medius; next-generation sequencing; qRT-PCR; viruses.
Figures
References
-
- Sazzad H. Nipah outbreak in Faridpur District, Bangladesh, 2010. Health Sci Bull. 2010;8:6–11.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

