Tomato fruit ripening factor NOR controls leaf senescence
- PMID: 31002305
- PMCID: PMC6506771
- DOI: 10.1093/jxb/erz098
Tomato fruit ripening factor NOR controls leaf senescence
Abstract
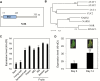
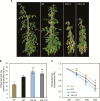
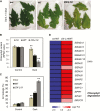
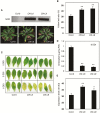
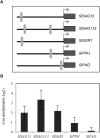
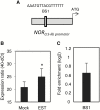
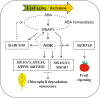
NAC transcription factors (TFs) are important regulators of expressional reprogramming during plant development, stress responses, and leaf senescence. NAC TFs also play important roles in fruit ripening. In tomato (Solanum lycopersicum), one of the best characterized NACs involved in fruit ripening is NON-RIPENING (NOR), and the non-ripening (nor) mutation has been widely used to extend fruit shelf life in elite varieties. Here, we show that NOR additionally controls leaf senescence. Expression of NOR increases with leaf age, and developmental as well as dark-induced senescence are delayed in the nor mutant, while overexpression of NOR promotes leaf senescence. Genes associated with chlorophyll degradation as well as senescence-associated genes (SAGs) show reduced and elevated expression, respectively, in nor mutants and NOR overexpressors. Overexpression of NOR also stimulates leaf senescence in Arabidopsis thaliana. In tomato, NOR supports senescence by directly and positively regulating the expression of several senescence-associated genes including, besides others, SlSAG15 and SlSAG113, SlSGR1, and SlYLS4. Finally, we find that another senescence control NAC TF, namely SlNAP2, acts upstream of NOR to regulate its expression. Our data support a model whereby NAC TFs have often been recruited by higher plants for both the control of leaf senescence and fruit ripening.
Keywords: Aging; NAC; NOR; leaf; non-ripening; senescence; tomato; transcription factor.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

Similar articles
-
NAC Transcription Factor Family Regulation of Fruit Ripening and Quality: A Review.Cells. 2022 Feb 2;11(3):525. doi: 10.3390/cells11030525. Cells. 2022. PMID: 35159333 Free PMC article. Review.
-
The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato.Plant Physiol. 2018 Jul;177(3):1286-1302. doi: 10.1104/pp.18.00292. Epub 2018 May 14. Plant Physiol. 2018. PMID: 29760199 Free PMC article.
-
A tomato NAC transcription factor, SlNAM1, positively regulates ethylene biosynthesis and the onset of tomato fruit ripening.Plant J. 2021 Dec;108(5):1317-1331. doi: 10.1111/tpj.15512. Epub 2021 Nov 2. Plant J. 2021. PMID: 34580960
-
The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review.Plant Mol Biol. 2021 Jun;106(3):223-238. doi: 10.1007/s11103-021-01128-w. Epub 2021 Feb 25. Plant Mol Biol. 2021. PMID: 33634368 Review.
-
A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation.Plant Cell Physiol. 2014 Jan;55(1):119-35. doi: 10.1093/pcp/pct162. Epub 2013 Nov 20. Plant Cell Physiol. 2014. PMID: 24265273
Cited by
-
NAC Transcription Factor Family Regulation of Fruit Ripening and Quality: A Review.Cells. 2022 Feb 2;11(3):525. doi: 10.3390/cells11030525. Cells. 2022. PMID: 35159333 Free PMC article. Review.
-
HEBE, a novel positive regulator of senescence in Solanum lycopersicum.Sci Rep. 2020 Jul 3;10(1):11021. doi: 10.1038/s41598-020-67937-z. Sci Rep. 2020. PMID: 32620827 Free PMC article.
-
Early transcriptional responses in Solanum peruvianum and Solanum lycopersicum account for different acclimation processes during water scarcity events.Sci Rep. 2021 Aug 5;11(1):15961. doi: 10.1038/s41598-021-95622-2. Sci Rep. 2021. PMID: 34354211 Free PMC article.
-
NACs, generalist in plant life.Plant Biotechnol J. 2023 Dec;21(12):2433-2457. doi: 10.1111/pbi.14161. Epub 2023 Aug 25. Plant Biotechnol J. 2023. PMID: 37623750 Free PMC article. Review.
-
Transcriptional regulation of tomato fruit ripening.Physiol Mol Biol Plants. 2024 Feb;30(2):289-303. doi: 10.1007/s12298-024-01424-x. Epub 2024 Mar 9. Physiol Mol Biol Plants. 2024. PMID: 38623160 Free PMC article. Review.
References
-
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. 2008. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology 10(Suppl 1), 63–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

