E6 proteins from high-risk HPV, low-risk HPV, and animal papillomaviruses activate the Wnt/β-catenin pathway through E6AP-dependent degradation of NHERF1
- PMID: 31002735
- PMCID: PMC6493770
- DOI: 10.1371/journal.ppat.1007575
E6 proteins from high-risk HPV, low-risk HPV, and animal papillomaviruses activate the Wnt/β-catenin pathway through E6AP-dependent degradation of NHERF1
Abstract
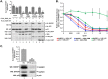
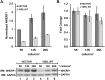
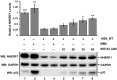
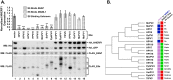
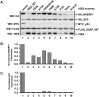
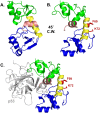
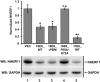
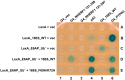
High-risk human papillomavirus (HPV) E6 proteins associate with the cellular ubiquitin ligase E6-Associated Protein (E6AP), and then recruit both p53 and certain cellular PDZ proteins for ubiquitination and degradation by the proteasome. Low-risk HPV E6 proteins also associate with E6AP, yet fail to recruit p53 or PDZ proteins; their E6AP-dependent targets have so far been uncharacterized. We found a cellular PDZ protein called Na+/H+ Exchanger Regulatory Factor 1 (NHERF1) is targeted for degradation by both high and low-risk HPV E6 proteins as well as E6 proteins from diverse non-primate mammalian species. NHERF1 was degraded by E6 in a manner dependent upon E6AP ubiquitin ligase activity but independent of PDZ interactions. A novel structural domain of E6, independent of the p53 recognition domain, was necessary to associate with and degrade NHERF1, and the NHERF1 EB domain was required for E6-mediated degradation. Degradation of NHERF1 by E6 activated canonical Wnt/β-catenin signaling, a key pathway that regulates cell growth and proliferation. Expression levels of NHERF1 increased with increasing cell confluency. This is the first study in which a cellular protein has been identified that is targeted for degradation by both high and low-risk HPV E6 as well as E6 proteins from diverse animal papillomaviruses. This suggests that NHERF1 plays a role in regulating squamous epithelial growth and further suggests that the interaction of E6 proteins with NHERF1 could be a common therapeutic target for multiple papillomavirus types.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Rector A, Van Ranst M. Animal papillomaviruses. J Virol. 2013;445(1):213–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

