Biogenesis, quality control, and structural dynamics of proteins as explored in living cells via site-directed photocrosslinking
- PMID: 31002747
- PMCID: PMC6566533
- DOI: 10.1002/pro.3627
Biogenesis, quality control, and structural dynamics of proteins as explored in living cells via site-directed photocrosslinking
Abstract
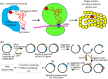
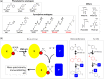
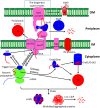
Protein biogenesis and quality control are essential to maintaining a functional pool of proteins and involve numerous protein factors that dynamically and transiently interact with each other and with the substrate proteins in living cells. Conventional methods are hardly effective for studying dynamic, transient, and weak protein-protein interactions that occur in cells. Herein, we review how the site-directed photocrosslinking approach, which relies on the genetic incorporation of a photoreactive unnatural amino acid into a protein of interest at selected individual amino acid residue positions and the covalent trapping of the interacting proteins upon ultraviolent irradiation, has become a highly efficient way to explore the aspects of protein contacts in living cells. For example, in the past decade, this approach has allowed the profiling of the in vivo substrate proteins of chaperones or proteases under both physiologically optimal and stressful (e.g., acidic) conditions, mapping residues located at protein interfaces, identifying new protein factors involved in the biogenesis of membrane proteins, trapping transiently formed protein complexes, and snapshotting different structural states of a protein. We anticipate that the site-directed photocrosslinking approach will play a fundamental role in dissecting the detailed mechanisms of protein biogenesis, quality control, and dynamics in the future.
Keywords: dynamics of proteins; in vivo protein photocrosslinking; membrane protein biogenesis; molecular chaperones; proteases; protein quality control.
© 2019 The Protein Society.
Figures

Similar articles
-
Photocrosslinking approaches to interactome mapping.Curr Opin Chem Biol. 2013 Feb;17(1):90-101. doi: 10.1016/j.cbpa.2012.10.034. Epub 2012 Nov 10. Curr Opin Chem Biol. 2013. PMID: 23149092 Free PMC article. Review.
-
Genetically encoded photocrosslinkers for identifying and mapping protein-protein interactions in living cells.IUBMB Life. 2016 Nov;68(11):879-886. doi: 10.1002/iub.1560. Epub 2016 Sep 26. IUBMB Life. 2016. PMID: 27670842 Review.
-
Engineering an acetyllysine reader with a photocrosslinking amino acid for interactome profiling.Chem Commun (Camb). 2021 Sep 28;57(77):9866-9869. doi: 10.1039/d1cc04611j. Chem Commun (Camb). 2021. PMID: 34490864
-
A photo-cross-linking approach to monitor folding and assembly of newly synthesized proteins in a living cell.J Biol Chem. 2018 Jan 12;293(2):677-686. doi: 10.1074/jbc.M117.817270. Epub 2017 Nov 20. J Biol Chem. 2018. PMID: 29158258 Free PMC article.
-
Genetically encoded releasable photo-cross-linking strategies for studying protein-protein interactions in living cells.Nat Protoc. 2017 Oct;12(10):2147-2168. doi: 10.1038/nprot.2017.090. Epub 2017 Sep 21. Nat Protoc. 2017. PMID: 28933779
Cited by
-
Visible Light-Induced Specific Protein Reaction Delineates Early Stages of Cell Adhesion.J Am Chem Soc. 2023 Nov 15;145(45):24459-24465. doi: 10.1021/jacs.3c07827. Epub 2023 Oct 31. J Am Chem Soc. 2023. PMID: 38104267 Free PMC article.
-
Visible light-induced specific protein reaction delineates early stages of cell adhesion.bioRxiv [Preprint]. 2023 Jul 22:2023.07.21.549850. doi: 10.1101/2023.07.21.549850. bioRxiv. 2023. Update in: J Am Chem Soc. 2023 Nov 15;145(45):24459-24465. doi: 10.1021/jacs.3c07827. PMID: 37503248 Free PMC article. Updated. Preprint.
-
Photocrosslinkable natural polymers in tissue engineering.Front Bioeng Biotechnol. 2023 Mar 2;11:1127757. doi: 10.3389/fbioe.2023.1127757. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 36970625 Free PMC article. Review.
-
Bifunctional Non-Canonical Amino Acids: Combining Photo-Crosslinking with Click Chemistry.Biomolecules. 2020 Apr 10;10(4):578. doi: 10.3390/biom10040578. Biomolecules. 2020. PMID: 32290035 Free PMC article. Review.
-
Non-Canonical Amino Acids in Analyses of Protease Structure and Function.Int J Mol Sci. 2023 Sep 13;24(18):14035. doi: 10.3390/ijms241814035. Int J Mol Sci. 2023. PMID: 37762340 Free PMC article. Review.
References
-
- Blobel G (2000) Protein targeting. ChemBioChem 1:86–102. - PubMed
-
- Chang Z. Biogenesis of secretory proteins: folding and quality control in the endoplasmic reticulum In: Bradshawa RA, Stahl P, Eds, 2016, Encyclopedia of cell biology. Waltham, MA: Academic Press; p. 535–544.
-
- Balchin D, Hayer‐Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353:aac4354. - PubMed
-
- Sontag EM, Samant RS, Frydman J (2017) Mechanisms and functions of spatial protein quality control. Annu Rev Biochem 86:97–122. - PubMed
-
- Chiti F, Dobson CM (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem 86:27–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

