Core Catalysis of the Reductive Glycine Pathway Demonstrated in Yeast
- PMID: 31002757
- PMCID: PMC6528164
- DOI: 10.1021/acssynbio.8b00464
Core Catalysis of the Reductive Glycine Pathway Demonstrated in Yeast
Abstract
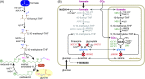
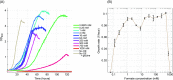
One-carbon (C1) compounds are attractive microbial feedstocks as they can be efficiently produced from widely available resources. Formate, in particular, represents a promising growth substrate, as it can be generated from electrochemical reduction of CO2 and fed to microorganisms in a soluble form. We previously identified the synthetic reductive glycine pathway as the most efficient route for aerobic growth on formate. We further demonstrated pathway activity in Escherichia coli after expression of both native and foreign genes. Here, we explore whether the reductive glycine pathway could be established in a model microorganism using only native enzymes. We used the yeast Saccharomyces cerevisiae as host and show that overexpression of only endogenous enzymes enables glycine biosynthesis from formate and CO2 in a strain that is otherwise auxotrophic for glycine. We find the pathway to be highly active in this host, where 0.125 mM formate is sufficient to support growth. Notably, the formate-dependent growth rate of the engineered S. cerevisiae strain remained roughly constant over a very wide range of formate concentrations, 1-500 mM, indicating both high affinity for formate use and high tolerance toward elevated concentration of this C1 feedstock. Our results, as well the availability of endogenous NAD-dependent formate dehydrogenase, indicate that yeast might be an especially suitable host for engineering growth on formate.
Keywords: carbon labeling; glycine cleavage system; metabolic engineering; one-carbon metabolism; synthetic biology; tetrahydrofolate.
Conflict of interest statement
The authors declare the following competing financial interest(s): A.B.E. is co-founder of b.fab, aiming to commercialize C1 assimilation. The company was not involved in any way in the conducting, funding, or influencing the research.
Figures


Similar articles
-
In Vivo Assimilation of One-Carbon via a Synthetic Reductive Glycine Pathway in Escherichia coli.ACS Synth Biol. 2018 Sep 21;7(9):2023-2028. doi: 10.1021/acssynbio.8b00131. Epub 2018 Jul 2. ACS Synth Biol. 2018. PMID: 29763299
-
Engineering and evolution of the complete Reductive Glycine Pathway in Saccharomyces cerevisiae for formate and CO2 assimilation.Metab Eng. 2024 Jan;81:167-181. doi: 10.1016/j.ymben.2023.11.007. Epub 2023 Nov 30. Metab Eng. 2024. PMID: 38040111
-
Paving the way for synthetic C1 - Metabolism in Pseudomonas putida through the reductive glycine pathway.Metab Eng. 2023 Mar;76:215-224. doi: 10.1016/j.ymben.2023.02.004. Epub 2023 Feb 15. Metab Eng. 2023. PMID: 36804222
-
Recent progress in metabolic engineering of microbial formate assimilation.Appl Microbiol Biotechnol. 2020 Aug;104(16):6905-6917. doi: 10.1007/s00253-020-10725-6. Epub 2020 Jun 21. Appl Microbiol Biotechnol. 2020. PMID: 32566995 Review.
-
Synthetic Methanol and Formate Assimilation Via Modular Engineering and Selection Strategies.Curr Issues Mol Biol. 2019;33:237-248. doi: 10.21775/cimb.033.237. Epub 2019 Jun 5. Curr Issues Mol Biol. 2019. PMID: 31166196 Review.
Cited by
-
Biocatalytic C-C Bond Formation for One Carbon Resource Utilization.Int J Mol Sci. 2021 Feb 14;22(4):1890. doi: 10.3390/ijms22041890. Int J Mol Sci. 2021. PMID: 33672882 Free PMC article. Review.
-
Enhancing xylanase expression by Komagataella phaffii by formate as carbon source and inducer.Appl Microbiol Biotechnol. 2022 Dec;106(23):7819-7829. doi: 10.1007/s00253-022-12249-7. Epub 2022 Oct 29. Appl Microbiol Biotechnol. 2022. PMID: 36307629
-
Perspectives for Using CO2 as a Feedstock for Biomanufacturing of Fuels and Chemicals.Bioengineering (Basel). 2023 Nov 26;10(12):1357. doi: 10.3390/bioengineering10121357. Bioengineering (Basel). 2023. PMID: 38135948 Free PMC article. Review.
-
The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans.Nat Commun. 2020 Oct 9;11(1):5090. doi: 10.1038/s41467-020-18906-7. Nat Commun. 2020. PMID: 33037220 Free PMC article.
-
Metabolic engineering strategies for microbial utilization of methanol.Eng Microbiol. 2023 Mar 4;3(3):100081. doi: 10.1016/j.engmic.2023.100081. eCollection 2023 Sep. Eng Microbiol. 2023. PMID: 39628934 Free PMC article. Review.
References
-
- Abubackar H. N.; eiga M. C.; Kennes C. (2011) Biological conversion of carbon monoxide: rich syngas or waste gases to bioethanol. Biofuels, Bioprod. Biorefin. 5, 93–114. 10.1002/bbb.256. - DOI
-
- Kopljar D.; Inan A.; Vindayer P.; Wagner N.; Klemm E. (2014) Electrochemical reduction of CO2 to formate at high current density using gas diffusion electrodes. J. Appl. Electrochem. 44, 1107–1116. 10.1007/s10800-014-0731-x. - DOI
-
- Yang H.; Kaczur J. J.; Sajjad S. D.; Masel R. I. (2017) Electrochemical conversion of CO2 to formic acid utilizing Sustainion membranes. Journal of CO2 Utilization 20, 208–217. 10.1016/j.jcou.2017.04.011. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

