SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species
- PMID: 31002797
- PMCID: PMC6499390
- DOI: 10.1016/j.cell.2019.03.043
SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species
Abstract
DNA repair has been hypothesized to be a longevity determinant, but the evidence for it is based largely on accelerated aging phenotypes of DNA repair mutants. Here, using a panel of 18 rodent species with diverse lifespans, we show that more robust DNA double-strand break (DSB) repair, but not nucleotide excision repair (NER), coevolves with longevity. Evolution of NER, unlike DSB, is shaped primarily by sunlight exposure. We further show that the capacity of the SIRT6 protein to promote DSB repair accounts for a major part of the variation in DSB repair efficacy between short- and long-lived species. We dissected the molecular differences between a weak (mouse) and a strong (beaver) SIRT6 protein and identified five amino acid residues that are fully responsible for their differential activities. Our findings demonstrate that DSB repair and SIRT6 have been optimized during the evolution of longevity, which provides new targets for anti-aging interventions.
Keywords: DNA DSB repair; DNA repair; NER; SIRT6; aging; longevity.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


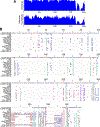


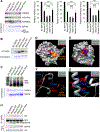

References
-
- Batista LF, Kaina B, Meneghini R, and Menck CF (2009). How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis. Mutat Res 681, 197–208. - PubMed
-
- Bodgi L, Canet A, Pujo-Menjouet L, Lesne A, Victor JM, and Foray N (2016). Mathematical models of radiation action on living cells: From the target theory to the modern approaches. A historical and critical review. J Theor Biol 394, 93–101. - PubMed
-
- Bovet J (1984). Strategies of Homing Behavior in the Red Squirrel, Tamiasciurus-Hudsonicus. Behav Ecol Sociobiol 16, 81–88.
-
- Bovet J, and Oertli EF (1974). Free-running circadian activity rhythms in free-living beaver (Castor canadensis). Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology 92, 1–10.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

