Genome-Wide Identification and Characterization of the OPR Gene Family in Wheat (Triticum aestivum L.)
- PMID: 31003470
- PMCID: PMC6514991
- DOI: 10.3390/ijms20081914
Genome-Wide Identification and Characterization of the OPR Gene Family in Wheat (Triticum aestivum L.)
Abstract
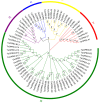
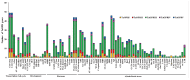
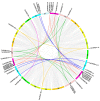
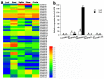
The 12-oxo-phytodienoic acid reductases (OPRs), which belong to the old yellow enzyme (OYE) family, are flavin mononucleotide (FMN)-dependent oxidoreductases with critical functions in plants. Despite the clear characteristics of growth and development, as well as the defense responses in Arabidopsis, tomato, rice, and maize, the potential roles of OPRs in wheat are not fully understood. Here, forty-eight putative OPR genes were found and classified into five subfamilies, with 6 in sub. I, 4 in sub. II, 33 in sub. III, 3 in sub. IV, and 2 in sub. V. Similar gene structures and conserved protein motifs of TaOPRs in wheat were identified in the same subfamilies. An analysis of cis-acting elements in promoters revealed that the functions of OPRs in wheat were mostly related to growth, development, hormones, biotic, and abiotic stresses. A total of 14 wheat OPR genes were identified as tandem duplicated genes, while 37 OPR genes were segmentally duplicated genes. The expression patterns of TaOPRs were tissue- and stress-specific, and the expression of TaOPRs could be regulated or induced by phytohormones and various stresses. Therefore, there were multiple wheat OPR genes, classified into five subfamilies, with functional diversification and specific expression patterns, and to our knowledge, this was the first study to systematically investigate the wheat OPR gene family. The findings not only provide a scientific foundation for the comprehensive understanding of the wheat OPR gene family, but could also be helpful for screening more candidate genes and breeding new varieties of wheat, with a high yield and stress resistance.
Keywords: OPR; expression pattern; gene family; stress response; wheat.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants.BMC Evol Biol. 2009 May 5;9:90. doi: 10.1186/1471-2148-9-90. BMC Evol Biol. 2009. PMID: 19416520 Free PMC article.
-
Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L.Int J Mol Sci. 2021 Jul 9;22(14):7396. doi: 10.3390/ijms22147396. Int J Mol Sci. 2021. PMID: 34299014 Free PMC article.
-
Genome-Wide Identification and Analysis of the NPR1-Like Gene Family in Bread Wheat and Its Relatives.Int J Mol Sci. 2019 Nov 27;20(23):5974. doi: 10.3390/ijms20235974. Int J Mol Sci. 2019. PMID: 31783558 Free PMC article.
-
Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): insights into the roles in biological processes especially stress response.Biometals. 2018 Dec;31(6):1019-1042. doi: 10.1007/s10534-018-0146-y. Epub 2018 Oct 4. Biometals. 2018. PMID: 30288657 Review.
-
The LONELY GUY gene family: from mosses to wheat, the key to the formation of active cytokinins in plants.Plant Biotechnol J. 2022 Apr;20(4):625-645. doi: 10.1111/pbi.13783. Epub 2022 Mar 1. Plant Biotechnol J. 2022. PMID: 35108444 Free PMC article. Review.
Cited by
-
Identification and Characterization of Histone Modification Gene Families and Their Expression Patterns During Pod and Seed Development in Peanut.Int J Mol Sci. 2025 Mar 13;26(6):2591. doi: 10.3390/ijms26062591. Int J Mol Sci. 2025. PMID: 40141232 Free PMC article.
-
Comparative analysis of VMT genes/proteins in selected plant species with emphasis on bread wheat (Triticum aestivum L.).Genes Genomics. 2023 Nov;45(11):1445-1461. doi: 10.1007/s13258-023-01427-0. Epub 2023 Jul 26. Genes Genomics. 2023. PMID: 37493927
-
Genome-wide analysis of OPR family genes in Vitis vinifera and the role of VvOPR1 in copper, zinc tolerance.Front Plant Sci. 2025 Feb 26;16:1509472. doi: 10.3389/fpls.2025.1509472. eCollection 2025. Front Plant Sci. 2025. PMID: 40078634 Free PMC article.
-
A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses.Int J Mol Sci. 2024 Jun 27;25(13):7054. doi: 10.3390/ijms25137054. Int J Mol Sci. 2024. PMID: 39000161 Free PMC article.
-
Genome-wide identification, gene cloning, subcellular location and expression analysis of the OPR gene family under salt stress in sweetpotato.BMC Plant Biol. 2024 Dec 6;24(1):1171. doi: 10.1186/s12870-024-05887-8. BMC Plant Biol. 2024. PMID: 39643880 Free PMC article.
References
-
- Savchenko T., Kolla V.A., Wang C.Q., Nasafi Z., Hicks D.R., Phadungchob B., Chehab W.E., Brandizzi F., Froehlich J., Dehesh K. Functional Convergence of Oxylipin and Abscisic Acid Pathways Controls Stomatal Closure in Response to Drought. Plant Physiol. 2014;164:1151–1160. doi: 10.1104/pp.113.234310. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

