Global Brain Transcriptome Analysis of a Tpp1 Neuronal Ceroid Lipofuscinoses Mouse Model
- PMID: 31003587
- PMCID: PMC6475859
- DOI: 10.1177/1759091419843393
Global Brain Transcriptome Analysis of a Tpp1 Neuronal Ceroid Lipofuscinoses Mouse Model
Abstract
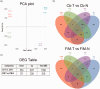
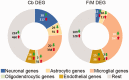
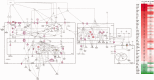
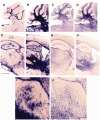
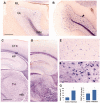
In humans, homozygous mutations in the TPP1 gene results in loss of tripeptidyl peptidase 1 (TPP1) enzymatic activity, leading to late infantile neuronal ceroid lipofuscinoses disease. Using a mouse model that targets the Tpp1 gene and recapitulates the pathology and clinical features of the human disease, we analyzed end-stage (4 months) transcriptional changes associated with lack of TPP1 activity. Using RNA sequencing technology, Tpp1 expression changes in the forebrain/midbrain and cerebellum of 4-month-old homozygotes were compared with strain-related controls. Transcriptional changes were found in 510 and 1,550 gene transcripts in forebrain/midbrain and cerebellum, respectively, from Tpp1-deficient brain tissues when compared with age-matched controls. Analysis of the differentially expressed genes using the Ingenuity™ pathway software, revealed increased neuroinflammation activity in microglia and astrocytes that could lead to neuronal dysfunction, particularly in the cerebellum. We also observed upregulation in the production of nitric oxide and reactive oxygen species; activation of leukocyte extravasation signals and complement pathways; and downregulation of major transcription factors involved in control of circadian rhythm. Several of these expression changes were confirmed by independent quantitative polymerase chain reaction and histological analysis by mRNA in situ hybridization, which allowed for an in-depth anatomical analysis of the pathology and provided independent confirmation of at least two of the major networks affected in this model. The identification of differentially expressed genes has revealed new lines of investigation for this complex disorder that may lead to novel therapeutic targets.
Keywords: circadian rhythm; lysosomal tripeptidyl peptidase 1; neuroinflammation; neuronal ceroid lipofuscinoses; pediatric neurodegeneration; transcriptome.
Figures




References
-
- Abreu T., Braganca , M. (2015). The bipolarity of light and dark: A review on bipolar disorder and circadian cycles. J Affect Disord, 185, 219–229. - PubMed
-
- Akiyama H., Nishimura T., Kondo H.Ikeda, K., Hayashi, Y., & McGeer, P. L. (1994). Expression of the receptor for macrophage colony stimulating factor by brain microglia and its upregulation in brains of patients with Alzheimer’s disease and amyotrophic lateral sclerosis. Brain Res, 639, 171–174. - PubMed
-
- Anderson G. W., Goebel H. H., Simonati , A. (2013). Human pathology in NCL. Biochim Biophys Acta, 1832, 1807–1826. - PubMed
-
- Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Bevan R. J., Evans R., Griffiths L.Watkins, L. M., Rees, M. I., Magliozzi, R., Allen, I., McDonnell, G., Kee, R., Naughton, M., Fitzgerald, D. C., Reynolds, R., Neal, J. W., & Howell, O. W. (2018). Meningeal inflammation and cortical demyelination in acute multiple sclerosis. Ann Neurol, 84, 829–842. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

