Deep learning only by normal brain PET identify unheralded brain anomalies
- PMID: 31003928
- PMCID: PMC6557913
- DOI: 10.1016/j.ebiom.2019.04.022
Deep learning only by normal brain PET identify unheralded brain anomalies
Abstract
Background: Recent deep learning models have shown remarkable accuracy for the diagnostic classification. However, they have limitations in clinical application due to the gap between the training cohorts and real-world data. We aimed to develop a model trained only by normal brain PET data with an unsupervised manner to identify an abnormality in various disorders as imaging data of the clinical routine.
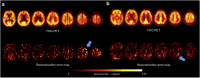
Methods: Using variational autoencoder, a type of unsupervised learning, Abnormality Score was defined as how far a given brain image is from the normal data. The model was applied to FDG PET data of Alzheimer's disease (AD) and mild cognitive impairment (MCI) and clinical routine FDG PET data for assessing behavioral abnormality and seizures. Accuracy was measured by the area under curve (AUC) of receiver-operating-characteristic (ROC) curve. We investigated whether deep learning has additional benefits with experts' visual interpretation to identify abnormal patterns.
Findings: The AUC of the ROC curve for differentiating AD was 0.90. The changes in cognitive scores from baseline to 2-year follow-up were significantly correlated with Abnormality Score at baseline. The AUC of the ROC curve for discriminating patients with various disorders from controls was 0.74. Experts' visual interpretation was helped by the deep learning model to identify abnormal patterns in 60% of cases initially not identified without the model.
Interpretation: We suggest that deep learning model trained only by normal data was applicable for identifying wide-range of abnormalities in brain diseases, even uncommon ones, proposing its possible use for interpreting real-world clinical data.
Keywords: Alzheimer; Anomaly detection; Deep learning; PET; Variational autoencoder.
Copyright © 2019. Published by Elsevier B.V.
Figures



References
-
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436. - PubMed
-
- Gulshan V., Peng L., Coram M. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama. 2016;316(22):2402–2410. - PubMed
-
- Choi H., Jin K.H., Initiative AsDN Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res. 2018;344:103–109. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

