Congo Red Fluorescence for Rapid In Situ Characterization of Synthetic Curli Systems
- PMID: 31003987
- PMCID: PMC6581178
- DOI: 10.1128/AEM.00434-19
Congo Red Fluorescence for Rapid In Situ Characterization of Synthetic Curli Systems
Abstract
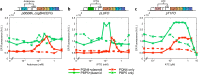
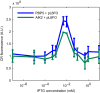
Curli are amyloid proteins that are assembled into extracellular polymeric fibers by bacteria during biofilm formation. The beta-sheet-rich protein CsgA, the primary structural component of the fibers, is secreted through dedicated machinery and self-assembles into cell-anchored fibers many times longer than the cell. Here, we have developed an in situ fluorescence assay for curli production that exploits the fluorescent properties of Congo red (CR) dye when bound to amyloid, allowing for rapid and robust curli quantification. We initially evaluated three amyloid-binding dyes for the fluorescent detection of curli in bacterial culture and found only Congo red compatible with in situ quantification. We further characterized the fluorescent properties of the dye directly in bacterial culture and calibrated the fluorescence using purified CsgA protein. We then used the Congo red assay to rapidly develop and characterize inducible curli-producing constructs in both an MC4100-derived lab strain of Escherichia coli and a derivative of the probiotic strain E. coli Nissle. This technique can be used to evaluate curli production in a minimally invasive manner using a range of equipment, simplifying curli quantification and the development of novel engineered curli systems.IMPORTANCE Curli are proteins produced by many bacteria as a structural component of biofilms, and they have recently emerged as a platform for fabrication of biological materials. Curli fibers are very robust and resistant to degradation, and the curli subunits can tolerate many protein fusions, facilitating the biosynthesis of novel functional materials. A serious bottleneck in the development of more sophisticated engineered curli systems is the rapid quantification of curli production by the bacteria. In this work we address this issue by developing a technique to monitor curli production directly in bacterial cultures, allowing for rapid curli quantification in a manner compatible with many powerful high-throughput techniques that can be used to engineer complex biological material systems.
Keywords: biofilms; biotechnology; curli; quantitative methods; synthetic biology.
Copyright © 2019 American Society for Microbiology.
Figures
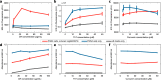
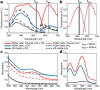

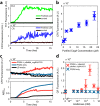
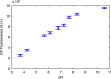
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

