Bacterial and Fungal Endophytic Microbiomes of Salicornia europaea
- PMID: 31003988
- PMCID: PMC6581177
- DOI: 10.1128/AEM.00305-19
Bacterial and Fungal Endophytic Microbiomes of Salicornia europaea
Abstract
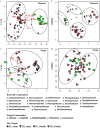
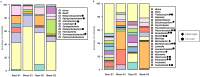
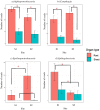
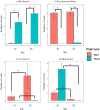
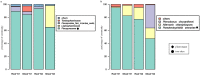
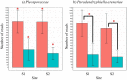
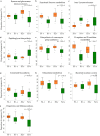
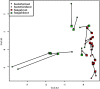
We examined Salicornia europaea, a nonmycorrhizal halophyte associated with specific and unique endophytic bacteria and fungi. The microbial community structure was analyzed at two sites differing in salinization history (anthropogenic and naturally saline site), in contrasting seasons (spring and fall) and in two plant organs (shoots and roots) via 16S rRNA and internal transcribed spacer amplicon sequencing. We observed distinct communities at the two sites, and in shoots and roots, while the season was of no importance. The bacterial community was less diverse in shoot libraries than in roots, regardless of the site and season, whereas no significant differences were observed for the fungal community. Proteobacteria and Bacteroidetes dominated bacterial assemblages, and Ascomycetes were the most frequent fungi. A root core microbiome operational taxonomic unit belonging to the genus Marinimicrobium was identified. We detected a significant influence of the Salicornia bacterial community on the fungal one by means of cocorrespondence analysis. In addition, pathways and potential functions of the bacterial community in Salicornia europaea were inferred and discussed. We can conclude that bacterial and fungal microbiomes of S. europaea are determined by the origin of salinity at the sites. Bacterial communities seemed to influence fungal ones, but not the other way around, which takes us closer to understanding of interactions between the two microbial groups. In addition, the plant organs of the halophyte filter the microbial community composition.IMPORTANCE Endophytes are particularly fascinating because of their multifaceted lifestyle, i.e., they may exist as either free-living soil microbes or saprobic ones or pathogens. Endophytic communities of halophytes may be different than those in other plants because salinity acts as an environmental filter. At the same time, they may contribute to the host's adaptation to adverse environmental conditions, which may be of importance in agriculture.
Keywords: 16S rRNA and ITS amplicon sequencing; endophyte; halophyte; microbial community structure; soil salinity.
Copyright © 2019 Furtado et al.
Figures

Similar articles
-
Bacterial microbiome of root-associated endophytes of Salicornia europaea in correspondence to different levels of salinity.Environ Sci Pollut Res Int. 2018 Sep;25(25):25420-25431. doi: 10.1007/s11356-018-2530-0. Epub 2018 Jun 27. Environ Sci Pollut Res Int. 2018. PMID: 29951760 Free PMC article.
-
High-Throughput Sequencing Analysis of the Endophytic Bacterial Diversity and Dynamics in Roots of the Halophyte Salicornia europaea.Curr Microbiol. 2016 May;72(5):557-62. doi: 10.1007/s00284-016-0990-3. Epub 2016 Jan 20. Curr Microbiol. 2016. PMID: 26787546
-
[Estimation of endophytic bacterial diversity in root of halophytes in Northern Xinjiang by high throughput sequencing].Wei Sheng Wu Xue Bao. 2016 Oct 4;56(10):1583-94. Wei Sheng Wu Xue Bao. 2016. PMID: 29741347 Chinese.
-
A Meta-Analysis Approach to Defining the Culturable Core of Plant Endophytic Bacterial Communities.Appl Environ Microbiol. 2022 Mar 22;88(6):e0253721. doi: 10.1128/aem.02537-21. Epub 2022 Feb 9. Appl Environ Microbiol. 2022. PMID: 35138928 Free PMC article. Review.
-
The contribution of beneficial wheat seed fungal communities beyond disease-causing fungi: Advancing heritable mycobiome-based plant breeding.Environ Microbiol Rep. 2024 Dec;16(6):e70004. doi: 10.1111/1758-2229.70004. Environ Microbiol Rep. 2024. PMID: 39529232 Free PMC article. Review.
Cited by
-
Endorhizosphere of indigenous succulent halophytes: a valuable resource of plant growth promoting bacteria.Environ Microbiome. 2023 Mar 18;18(1):20. doi: 10.1186/s40793-023-00477-x. Environ Microbiome. 2023. PMID: 36934265 Free PMC article.
-
The anticancer and antibacterial potential of bioactive secondary metabolites derived From bacterial endophytes in association with Artemisia absinthium.Sci Rep. 2023 Oct 27;13(1):18473. doi: 10.1038/s41598-023-45910-w. Sci Rep. 2023. PMID: 37891400 Free PMC article.
-
When Salt Meddles Between Plant, Soil, and Microorganisms.Front Plant Sci. 2020 Sep 16;11:553087. doi: 10.3389/fpls.2020.553087. eCollection 2020. Front Plant Sci. 2020. PMID: 33042180 Free PMC article. Review.
-
Bacterial community structure of the sunflower (Helianthus annuus) endosphere.Plant Signal Behav. 2021 Dec 2;16(12):1974217. doi: 10.1080/15592324.2021.1974217. Epub 2021 Sep 30. Plant Signal Behav. 2021. PMID: 34590546 Free PMC article.
-
Deciphering the mechanisms, hormonal signaling, and potential applications of endophytic microbes to mediate stress tolerance in medicinal plants.Front Plant Sci. 2023 Nov 15;14:1250020. doi: 10.3389/fpls.2023.1250020. eCollection 2023. Front Plant Sci. 2023. PMID: 38034581 Free PMC article. Review.
References
-
- Ushakova SA, Kovaleva NP, Gribovskaya IV, Dolgushev VA, Tikhomirova NA. 2005. Effect of NaCl concentration on productivity and mineral composition of Salicornia europaea as a potential crop for utilization NaCl in LSS. Adv Sp Res 36:1349–1353. doi:10.1016/j.asr.2004.09.017. - DOI
-
- Oliveira V, Gomes NCM, Cleary DFR, Almeida A, Silva AMS, Simões MMQ, Silva H, Cunha Â. 2014. Halophyte plant colonization as a driver of the composition of bacterial communities in salt marshes chronically exposed to oil hydrocarbons. FEMS Microbiol Ecol 90:647–662. doi:10.1111/1574-6941.12425. - DOI - PubMed
-
- Smillie C. 2015. Salicornia spp. as a biomonitor of Cu and Zn in salt marsh sediments. Ecol Indic 56:70–78. doi:10.1016/j.ecolind.2015.03.010. - DOI
-
- Abrol YP, Wild A. 2004. Soils, land, and food: managing the land during the twenty-first century. Ann Bot 93:785–786. doi:10.1093/aob/mch104. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

