Establishment Limitation Constrains the Abundance of Lactic Acid Bacteria in the Napa Cabbage Phyllosphere
- PMID: 31003989
- PMCID: PMC6581170
- DOI: 10.1128/AEM.00269-19
Establishment Limitation Constrains the Abundance of Lactic Acid Bacteria in the Napa Cabbage Phyllosphere
Abstract
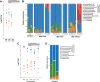
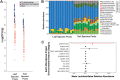
Patterns of phyllosphere diversity have become increasingly clear with high-throughput sequencing surveys, but the processes that control phyllosphere diversity are still emerging. Through a combination of lab and field experiments using Napa cabbage and lactic acid bacteria (LAB), we examined how dispersal and establishment processes shape the ecological distributions of phyllosphere bacteria. We first determined the abundance and diversity of LAB on Napa cabbage grown at three sites using both culture-based approaches and 16S rRNA gene amplicon sequencing. Across all sites, LAB made up less than 0.9% of the total bacterial community abundance. To assess whether LAB were low in abundance in the Napa cabbage phyllosphere due to a limited abundance in local species pools (source limitation), we quantified LAB in leaf and soil samples across 51 vegetable farms and gardens throughout the northeastern United States. Across all sites, LAB comprised less than 3.2% of the soil bacterial communities and less than 1.6% of phyllosphere bacterial communities. To assess whether LAB are unable to grow in the phyllosphere even if they dispersed at high rates (establishment limitation), we used a gnotobiotic Napa cabbage system in the lab with experimental communities mimicking various dispersal rates of LAB. Even at high dispersal rates, LAB became rare or completely undetectable in experimental communities, suggesting that they are also establishment limited. Collectively, our data demonstrate that the low abundance of LAB in phyllosphere communities may be explained by establishment limitation.IMPORTANCE The quality and safety of vegetable fermentations are dependent on the activities of LAB naturally present in the phyllosphere. Despite their critical role in determining the success of fermentation, the processes that determine the abundance and diversity of LAB in vegetables used for fermentation are poorly characterized. Our work demonstrates that the limited ability of LAB to grow in the cabbage phyllosphere environment may constrain their abundance on cabbage leaves. These results suggest that commercial fermentation of Napa cabbage proceeds despite low and variable abundances of LAB across different growing regions. Propagule limitation may also explain ecological distributions of other rare members of phyllosphere microbes.
Keywords: 16S; cabbage; community assembly; lactic acid bacteria; microbiome; phyllosphere.
Copyright © 2019 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

