CENTRORADIALIS Interacts with FLOWERING LOCUS T-Like Genes to Control Floret Development and Grain Number
- PMID: 31004004
- PMCID: PMC6548242
- DOI: 10.1104/pp.18.01454
CENTRORADIALIS Interacts with FLOWERING LOCUS T-Like Genes to Control Floret Development and Grain Number
Abstract
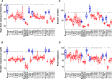
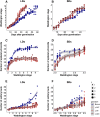
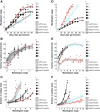
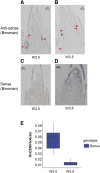
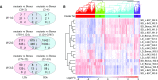
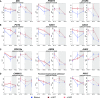
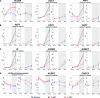
CENTRORADIALIS (CEN) is a key regulator of flowering time and inflorescence architecture in plants. Natural variation in the barley (Hordeum vulgare) homolog HvCEN is important for agricultural range expansion of barley cultivation, but its effects on shoot and spike architecture and consequently yield have not yet been characterized. Here, we evaluated 23 independent hvcen, also termed mat-c, mutants to determine the pleiotropic effects of HvCEN on developmental timing and shoot and spike morphologies of barley under outdoor and controlled conditions. All hvcen mutants flowered early and showed a reduction in spikelet number per spike, tiller number, and yield in the outdoor experiments. Mutations in hvcen accelerated spikelet initiation and reduced axillary bud number in a photoperiod-independent manner but promoted floret development only under long days (LDs). The analysis of a flowering locus t3 (hvft3) hvcen double mutant showed that HvCEN interacts with HvFT3 to control spikelet initiation. Furthermore, early flowering3 (hvelf3) hvcen double mutants with high HvFT1 expression levels under short days suggested that HvCEN interacts with HvFT1 to repress floral development. Global transcriptome profiling in developing shoot apices and inflorescences of mutant and wild-type plants revealed that HvCEN controlled transcripts involved in chromatin remodeling activities, cytokinin and cell cycle regulation and cellular respiration under LDs and short days, whereas HvCEN affected floral homeotic genes only under LDs. Understanding the stage and organ-specific functions of HvCEN and downstream molecular networks will allow the manipulation of different shoot and spike traits and thereby yield.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures

References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 - PubMed
-
- Alqudah AM, Schnurbusch T (2014) Awn primordium to tipping is the most decisive developmental phase for spikelet survival in barley. Funct Plant Biol 41: 424–436 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

