MAPK pathway and B cells overactivation in multiple sclerosis revealed by phosphoproteomics and genomic analysis
- PMID: 31004050
- PMCID: PMC6510997
- DOI: 10.1073/pnas.1818347116
MAPK pathway and B cells overactivation in multiple sclerosis revealed by phosphoproteomics and genomic analysis
Abstract
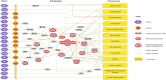
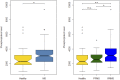
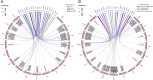
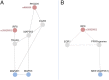
Dysregulation of signaling pathways in multiple sclerosis (MS) can be analyzed by phosphoproteomics in peripheral blood mononuclear cells (PBMCs). We performed in vitro kinetic assays on PBMCs in 195 MS patients and 60 matched controls and quantified the phosphorylation of 17 kinases using xMAP assays. Phosphoprotein levels were tested for association with genetic susceptibility by typing 112 single-nucleotide polymorphisms (SNPs) associated with MS susceptibility. We found increased phosphorylation of MP2K1 in MS patients relative to the controls. Moreover, we identified one SNP located in the PHDGH gene and another on IRF8 gene that were associated with MP2K1 phosphorylation levels, providing a first clue on how this MS risk gene may act. The analyses in patients treated with disease-modifying drugs identified the phosphorylation of each receptor's downstream kinases. Finally, using flow cytometry, we detected in MS patients increased STAT1, STAT3, TF65, and HSPB1 phosphorylation in CD19+ cells. These findings indicate the activation of cell survival and proliferation (MAPK), and proinflammatory (STAT) pathways in the immune cells of MS patients, primarily in B cells. The changes in the activation of these kinases suggest that these pathways may represent therapeutic targets for modulation by kinase inhibitors.
Keywords: B cells; autoimmunity; multiple sclerosis; phosphoproteomics; signaling pathways.
Conflict of interest statement
Conflict of interest statement: D.M. is an employee of ProtATonce; T.O. received honoraria for lectures and/or participation on advisory boards, as well as unrestricted multiple sclerosis research grants from Allmiral, Astrazeneca, Biogen, Genzyme, Merck, and Novartis; R.M. received grants and personal fees from Biogen Idec, personal fees from Genzyme Sanofi Aventis, grants and personal fees from Novartis, and personal fees from Merck Serono, Bionamics, all for work unrelated to that submitted; R.M. received honoraria for lectures and/or participation on advisory boards, as well as unrestricted multiple sclerosis research grants from Biogen, Genzyme, Merck, Celgene, Roche, Novartis, Neuway, and CellProtect, all for work unrelated to that submitted; F.P. received research grants and personal compensation from Alexion, Bayer, Chugai, Novartis, Merck, Teva, Sanofi, Genzyme, Biogen, and MedImmune; L.G.A is the founder and shareholder at ProtATonce; P.V. holds stock in and has received consultancy payments from Bionure Farma SL, QMenta Inc, Health Engineering SL, Spire Therapeutics Inc, and Spire Bioventures Inc.
Figures
Similar articles
-
Prediction of combination therapies based on topological modeling of the immune signaling network in multiple sclerosis.Genome Med. 2021 Jul 16;13(1):117. doi: 10.1186/s13073-021-00925-8. Genome Med. 2021. PMID: 34271980 Free PMC article.
-
Aberrant STAT phosphorylation signaling in peripheral blood mononuclear cells from multiple sclerosis patients.J Neuroinflammation. 2018 Mar 7;15(1):72. doi: 10.1186/s12974-018-1105-9. J Neuroinflammation. 2018. PMID: 29514694 Free PMC article.
-
Phosphoproteome analysis of the MAPK pathway reveals previously undetected feedback mechanisms.Proteomics. 2016 Jul;16(14):1998-2004. doi: 10.1002/pmic.201600119. Proteomics. 2016. PMID: 27273156
-
Comprehensive analysis of kinase-oriented phospho-signalling pathways.J Biochem. 2019 Apr 1;165(4):301-307. doi: 10.1093/jb/mvy115. J Biochem. 2019. PMID: 30590682 Review.
-
From Phosphosites to Kinases.Methods Mol Biol. 2016;1355:307-21. doi: 10.1007/978-1-4939-3049-4_21. Methods Mol Biol. 2016. PMID: 26584935 Review.
Cited by
-
VPS34-dependent control of apical membrane function of proximal tubule cells and nutrient recovery by the kidney.Sci Signal. 2022 Nov 29;15(762):eabo7940. doi: 10.1126/scisignal.abo7940. Epub 2022 Nov 29. Sci Signal. 2022. PMID: 36445937 Free PMC article.
-
Predicting disease severity in multiple sclerosis using multimodal data and machine learning.J Neurol. 2024 Mar;271(3):1133-1149. doi: 10.1007/s00415-023-12132-z. Epub 2023 Dec 22. J Neurol. 2024. PMID: 38133801 Free PMC article.
-
Multiscale networks in multiple sclerosis.PLoS Comput Biol. 2024 Feb 8;20(2):e1010980. doi: 10.1371/journal.pcbi.1010980. eCollection 2024 Feb. PLoS Comput Biol. 2024. PMID: 38329927 Free PMC article.
-
Quantitative proteomics and multi-omics analysis identifies potential biomarkers and the underlying pathological molecular networks in Chinese patients with multiple sclerosis.BMC Neurol. 2024 Oct 31;24(1):423. doi: 10.1186/s12883-024-03926-3. BMC Neurol. 2024. PMID: 39478468 Free PMC article.
-
Coupling the Paternò-Büchi (PB) Reaction With Mass Spectrometry to Study Unsaturated Fatty Acids in Mouse Model of Multiple Sclerosis.Front Chem. 2019 Nov 26;7:807. doi: 10.3389/fchem.2019.00807. eCollection 2019. Front Chem. 2019. PMID: 31850304 Free PMC article.
References
-
- Kotelnikova E, et al. Signaling networks in MS: A systems-based approach to developing new pharmacological therapies. Mult Scler. 2015;21:138–146. - PubMed
-
- Stasyk T, Huber LA. Mapping in vivo signal transduction defects by phosphoproteomics. Trends Mol Med. 2012;18:43–51. - PubMed
-
- Morris MK, Chi A, Melas IN, Alexopoulos LG. Phosphoproteomics in drug discovery. Drug Discov Today. 2014;19:425–432. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

