Component of splicing factor SF3b plays a key role in translational control of polyribosomes on the endoplasmic reticulum
- PMID: 31004060
- PMCID: PMC6510990
- DOI: 10.1073/pnas.1901742116
Component of splicing factor SF3b plays a key role in translational control of polyribosomes on the endoplasmic reticulum
Abstract
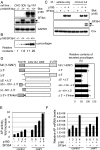
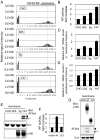
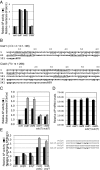
One of the morphological hallmarks of terminally differentiated secretory cells is highly proliferated membrane of the rough endoplasmic reticulum (ER), but the molecular basis for the high rate of protein biosynthesis in these cells remains poorly documented. An important aspect of ER translational control is the molecular mechanism that supports efficient use of targeted mRNAs in polyribosomes. Here, we identify an enhancement system for ER translation promoted by p180, an integral ER membrane protein we previously reported as an essential factor for the assembly of ER polyribosomes. We provide evidence that association of target mRNAs with p180 is critical for efficient translation, and that SF3b4, an RNA-binding protein in the splicing factor SF3b, functions as a cofactor for p180 at the ER and plays a key role in enhanced translation of secretory proteins. A cis-element in the 5' untranslated region of collagen and fibronectin genes is important to increase translational efficiency in the presence of p180 and SF3b4. These data demonstrate that a unique system comprising a p180-SF3b4-mRNA complex facilitates the selective assembly of polyribosomes on the ER.
Keywords: 5′ UTR; SF3b4; endoplasmic reticulum; p180; polyribosome.
Conflict of interest statement
Conflict of interest statement: T.U., Y.T., and K.O.-G. are coinventors of patent applications based on this work. T.U., Y.T., S.H., and K.O.-G. are employees of Nippi, Inc., an applicant of the patents (JP58488 and other related patent applications, including in the European Union and the United States). Nippi, Inc. is going to develop application of a new engineering technology (spERt Technology) based on ideas described in this study, which is likely to up-regulate productivity of recombinant proteins. Our development of spERt Technology has not influenced any conclusions of this study.
Figures



Similar articles
-
Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, p180.Nucleic Acids Res. 2012 Apr;40(7):3006-17. doi: 10.1093/nar/gkr1197. Epub 2011 Dec 7. Nucleic Acids Res. 2012. PMID: 22156060 Free PMC article.
-
Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion.J Biol Chem. 2010 Sep 24;285(39):29941-50. doi: 10.1074/jbc.M109.094607. Epub 2010 Jul 20. J Biol Chem. 2010. PMID: 20647306 Free PMC article.
-
p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum.PLoS Biol. 2012;10(5):e1001336. doi: 10.1371/journal.pbio.1001336. Epub 2012 May 29. PLoS Biol. 2012. PMID: 22679391 Free PMC article.
-
Localization of mRNAs to the endoplasmic reticulum.Wiley Interdiscip Rev RNA. 2014 Jul-Aug;5(4):481-92. doi: 10.1002/wrna.1225. Epub 2014 Mar 18. Wiley Interdiscip Rev RNA. 2014. PMID: 24644132 Review.
-
In vitro and tissue culture methods for analysis of translation initiation on the endoplasmic reticulum.Methods Enzymol. 2007;431:47-60. doi: 10.1016/S0076-6879(07)31004-5. Methods Enzymol. 2007. PMID: 17923230 Review.
Cited by
-
A dual role of RBM42 in modulating splicing and translation of CDKN1A/p21 during DNA damage response.Nat Commun. 2023 Nov 22;14(1):7628. doi: 10.1038/s41467-023-43495-6. Nat Commun. 2023. PMID: 37993446 Free PMC article.
-
Regulating Divergent Transcriptomes through mRNA Splicing and Its Modulation Using Various Small Compounds.Int J Mol Sci. 2020 Mar 16;21(6):2026. doi: 10.3390/ijms21062026. Int J Mol Sci. 2020. PMID: 32188117 Free PMC article. Review.
-
Multipurpose RNA maturation factors dysregulate multiple mRNA processing steps simultaneously and provide new therapeutic opportunities.RNA Biol. 2025 Dec;22(1):1-14. doi: 10.1080/15476286.2025.2503040. Epub 2025 Jun 9. RNA Biol. 2025. PMID: 40485569 Free PMC article. Review.
-
Exploration of the association between SF3B4 and HMGB1 expression and the clinicopathological features and prognosis of gastric cancer.World J Gastrointest Oncol. 2025 Aug 15;17(8):109120. doi: 10.4251/wjgo.v17.i8.109120. World J Gastrointest Oncol. 2025. PMID: 40837768 Free PMC article.
-
Long-read subcellular fractionation and sequencing reveals the translational fate of full-length mRNA isoforms during neuronal differentiation.Genome Res. 2024 Nov 20;34(11):2000-2011. doi: 10.1101/gr.279170.124. Genome Res. 2024. PMID: 38839373 Free PMC article.
References
-
- Palade G, editor. A Small Particulate Component of the Cytoplasm. Yale Univ Press; New Haven, CT: 1958. pp. 283–304.
-
- Christensen AK, Bourne CM. Shape of large bound polysomes in cultured fibroblasts and thyroid epithelial cells. Anat Rec. 1999;255:116–129. - PubMed
-
- Hermesh O, Jansen RP. Take the (RN)A-train: Localization of mRNA to the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2519–2525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

