Nudt8 is a novel CoA diphosphohydrolase that resides in the mitochondria
- PMID: 31004344
- PMCID: PMC6557688
- DOI: 10.1002/1873-3468.13392
Nudt8 is a novel CoA diphosphohydrolase that resides in the mitochondria
Abstract
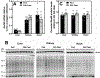
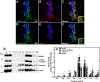
CoA regulates energy metabolism and exists in separate pools in the cytosol, peroxisomes, and mitochondria. At the whole tissue level, the concentration of CoA changes with the nutritional state by balancing synthesis and degradation; however, it is currently unclear how individual subcellular CoA pools are regulated. Liver and kidney peroxisomes contain Nudt7 and Nudt19, respectively, enzymes that catalyze CoA degradation. We report that Nudt8 is a novel CoA-degrading enzyme that resides in the mitochondria. Nudt8 has a distinctive preference for manganese ions and exhibits a broader tissue distribution than Nudt7 and Nudt19. The existence of CoA-degrading enzymes in both peroxisomes and mitochondria suggests that degradation may be a key regulatory mechanism for modulating the intracellular CoA pools.
Keywords: Coenzyme A; Nudix hydrolase; cell compartmentalization; metabolic regulation; mitochondria.
© 2019 Federation of European Biochemical Societies.
Figures

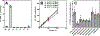

Similar articles
-
Regulation of coenzyme A levels by degradation: the 'Ins and Outs'.Prog Lipid Res. 2020 Apr;78:101028. doi: 10.1016/j.plipres.2020.101028. Epub 2020 Mar 29. Prog Lipid Res. 2020. PMID: 32234503 Free PMC article. Review.
-
Nudt19 is a renal CoA diphosphohydrolase with biochemical and regulatory properties that are distinct from the hepatic Nudt7 isoform.J Biol Chem. 2018 Mar 16;293(11):4134-4148. doi: 10.1074/jbc.RA117.001358. Epub 2018 Jan 29. J Biol Chem. 2018. PMID: 29378847 Free PMC article.
-
Overexpression of Nudt7 decreases bile acid levels and peroxisomal fatty acid oxidation in the liver.J Lipid Res. 2019 May;60(5):1005-1019. doi: 10.1194/jlr.M092676. Epub 2019 Mar 7. J Lipid Res. 2019. PMID: 30846528 Free PMC article.
-
The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives.Biochem J. 2001 Jul 1;357(Pt 1):33-8. doi: 10.1042/0264-6021:3570033. Biochem J. 2001. PMID: 11415433 Free PMC article.
-
Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes.Biochimie. 2014 Mar;98:45-55. doi: 10.1016/j.biochi.2013.12.018. Epub 2014 Jan 2. Biochimie. 2014. PMID: 24389458 Review.
Cited by
-
FIT2 is an acyl-coenzyme A diphosphatase crucial for endoplasmic reticulum homeostasis.J Cell Biol. 2020 Oct 5;219(10):e202006111. doi: 10.1083/jcb.202006111. J Cell Biol. 2020. PMID: 32915949 Free PMC article.
-
Regulation of coenzyme A levels by degradation: the 'Ins and Outs'.Prog Lipid Res. 2020 Apr;78:101028. doi: 10.1016/j.plipres.2020.101028. Epub 2020 Mar 29. Prog Lipid Res. 2020. PMID: 32234503 Free PMC article. Review.
-
Coenzyme A biosynthesis: mechanisms of regulation, function and disease.Nat Metab. 2024 Jun;6(6):1008-1023. doi: 10.1038/s42255-024-01059-y. Epub 2024 Jun 13. Nat Metab. 2024. PMID: 38871981 Review.
-
Thiophosphate Analogs of Coenzyme A and Its Precursors-Synthesis, Stability, and Biomimetic Potential.Biomolecules. 2022 Aug 1;12(8):1065. doi: 10.3390/biom12081065. Biomolecules. 2022. PMID: 36008959 Free PMC article.
-
Seed Germination in Oil Palm (Elaeis guineensis Jacq.): A Review of Metabolic Pathways and Control Mechanisms.Int J Mol Sci. 2020 Jun 13;21(12):4227. doi: 10.3390/ijms21124227. Int J Mol Sci. 2020. PMID: 32545810 Free PMC article. Review.
References
-
- Choudhary C, Weinert BT, Nishida Y, Verdin E, and Mann M (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nature reviews. Molecular cell biology 15, 536–550 - PubMed
-
- Daniotti JL, Pedro MP, and Valdez Taubas J. (2017) The role of S-acylation in protein trafficking. Traffic - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

