The frequency of CNVs in a cohort population of consecutive fetuses with congenital anomalies after the termination of pregnancy
- PMID: 31004418
- PMCID: PMC6565594
- DOI: 10.1002/mgg3.658
The frequency of CNVs in a cohort population of consecutive fetuses with congenital anomalies after the termination of pregnancy
Abstract
Background: The implementation of molecular karyotyping has resulted in an improved diagnostic yield in the genetic diagnostics of congenital anomalies, detected prenatally or after the termination of pregnancy. However, the systematic epidemiologic ascertainment of copy number variations in the etiology of congenital anomalies has not yet been sufficiently explored.
Methods: Consecutive fetuses, altogether 204, with major single or multiple congenital anomalies were ascertained by using the SLOCAT registry for the period from 2011 to 2015. After excluding aneuploidies by using conventional karyotyping or Quantitative Fluorescence-Polymerase Chain Reaction, array comparative genomic hybridization was performed for the detection of copy number variations.
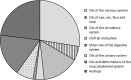
Results: We identified pathogenic or likely pathogenic copy number variations in 14 fetuses (6.8%); 2.9% in fetuses with isolated, and 3.9% in fetuses with multiple congenital anomalies. Additionally, aneuploidies and major structural chromosomal abnormalities were detected in 40.2%.
Conclusion: Our systematic approach of ascertaining congenital anomalies resulted in explaining the etiology of congenital anomalies in 47% of fetuses after the termination of pregnancy. By using array comparative genomic hybridization, we found that copy number variations represent an important part in the etiology of multiple, as well as isolated congenital anomalies, which indicates the importance of analyzing copy number variations in the diagnostic approach of fetuses with congenital anomalies after the termination of pregnancy.
Keywords: congenital anomalies; copy number variations; epidemiology; molecular karyotyping; termination of pregnancy.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
This research received no specific grant from any funding agency in the public, commercial, or not‐for‐profit sectors. The authors stated that there are no conflicts of interest regarding the publication of this manuscript. The authors also state that they have had full control of all primary data and that they agree to allow the Journal to review their data if requested.
Figures
References
-
- Béna, F. , Bruno, D. L. , Eriksson, M. , van Ravenswaaij‐Arts, C. , Stark, Z. , Dijkhuizen, T. , … Schoumans, J. (2013). Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics., 162B, 388–403. 10.1002/ajmg.b.32148 - DOI - PubMed
-
- D'Amours, G. , Kibar, Z. , Mathonnet, G. , Fetni, R. , Tihy, F. , Désilets, V. , … Lemyre, E. (2012). Whole‐genome array CGH identifies pathogenic copy number variations in fetuses with major malformations and a normal karyotype. Clinical Genetics, 81, 128–141. 10.1111/j.1399-0004.2011.01687.x - DOI - PubMed
-
- Di Gregorio, E. , Gai, G. , Botta, G. , Calcia, A. , Pappi, P. , Talarico, F. , … Brussino, A. (2015). Array‐comparative genomic hybridization analysis in fetuses with major congenital malformations reveals that 24% of cases have pathogenic deletions/duplications. Cytogenet Genome Research, 147, 10–16. 10.1159/000442308 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical