Using a hybrid read-across method to evaluate chemical toxicity based on chemical structure and biological data
- PMID: 31004930
- PMCID: PMC6508079
- DOI: 10.1016/j.ecoenv.2019.04.019
Using a hybrid read-across method to evaluate chemical toxicity based on chemical structure and biological data
Abstract
Read-across has become a primary approach to fill data gaps for chemical safety assessments. Chemical similarity based on structure, reactivity, and physic-chemical property information is a traditional approach applied for read-across toxicity studies. However, toxicity mechanisms are usually complicated in a biological system, so only using chemical similarity to perform the read-across for new compounds was not satisfactory for most toxicity endpoints, especially when the chemically similar compounds show dissimilar toxicities. This study aims to develop an enhanced read-across method for chemical toxicity predictions. To this end, we used two large toxicity datasets for read-across purposes. One consists of 3979 compounds with Ames mutagenicity data, and the other contains 7332 compounds with rat acute oral toxicity data. First, biological data for all compounds in these two datasets were obtained by querying thousands of PubChem bioassays. The PubChem bioassays with at least five compounds from either of these two datasets showing active responses were selected to generate comprehensive bioprofiles. The read-across studies were performed by using chemical similarity search only and also by using a hybrid similarity search based on both chemical descriptors and bioprofiles. Compared to traditional read-across based on chemical similarity, the hybrid read-across approach showed improved accuracy of predictions for both Ames mutagenicity and acute oral toxicity. Furthermore, we could illustrate potential toxicity mechanisms by analyzing the bioprofiles used for this hybrid read-across study. The results of this study indicate that the new hybrid read-across approach could be an applicable computational tool for chemical toxicity predictions. In this way, the bottleneck of traditional read-across studies can be overcome by introducing public biological data into the traditional process. The incorporation of bioprofiles generated from the additional biological data for compounds can partially solve the "activity cliff" issue and reveal their potential toxicity mechanisms. This study leads to a promising direction to utilize data-driven approaches for computational toxicology studies in the big data era.
Keywords: Big data; Biosimilarity; Computational toxicology; Hybrid approach; Read-across; Toxicity mechanisms.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure statement
No potential conflict of interest was reported by the authors.
Figures
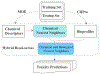
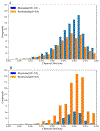
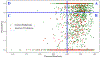
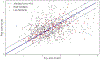
References
-
- Bakhtyari NG, Raitano G, Benfenati E, Martin T, Young D. 2013. Comparison of in silico models for prediction of mutagenicity. Journal of Environmental Science and Health, Part C 31:45–66. - PubMed
-
- Balls M 1994. Replacement of animal procedures: Alternatives in research, education and testing. Lab Anim 28:193–211. - PubMed
-
- Baumans V 2004. Use of animals in experimental research: An ethical dilemma? Gene Ther 11 Suppl 1:S64–66. - PubMed
-
- Belenky P, Bogan KL, Brenner C. 2007. Nad+ metabolism in health and disease. Trends in Biochemical Sciences 32:12–19. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

