Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation
- PMID: 31004987
- PMCID: PMC7125799
- DOI: 10.1016/j.virol.2019.03.017
Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation
Abstract
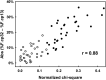
Overlapping genes represent an intriguing puzzle, as they encode two proteins whose ability to evolve is constrained by each other. Overlapping genes can undergo "symmetric evolution" (similar selection pressures on the two proteins) or "asymmetric evolution" (significantly different selection pressures on the two proteins). By sequence analysis of 75 pairs of homologous viral overlapping genes, I evaluated their accordance with one or the other model. Analysis of nucleotide and amino acid sequences revealed that half of overlaps undergo asymmetric evolution, as the protein from one frame shows a number of substitutions significantly higher than that of the protein from the other frame. Interestingly, the most variable protein (often known to interact with the host proteins) appeared to be encoded by the de novo frame in all cases examined. These findings suggest that overlapping genes, besides to increase the coding ability of viruses, are also a source of selective protein adaptation.
Keywords: Ancestral frame; De novo frame; Homologs; Non-synonymous nucleotide substitution; Selection pressure; Synonymous nucleotide substitution; Virus adaptation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
None.
Figures
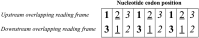
Similar articles
-
New insights into the evolutionary features of viral overlapping genes by discriminant analysis.Virology. 2020 Jul;546:51-66. doi: 10.1016/j.virol.2020.03.007. Epub 2020 Apr 2. Virology. 2020. PMID: 32452417 Free PMC article.
-
Origin, Evolution and Stability of Overlapping Genes in Viruses: A Systematic Review.Genes (Basel). 2021 May 26;12(6):809. doi: 10.3390/genes12060809. Genes (Basel). 2021. PMID: 34073395 Free PMC article.
-
Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation.J Virol. 2009 Oct;83(20):10719-36. doi: 10.1128/JVI.00595-09. Epub 2009 Jul 29. J Virol. 2009. PMID: 19640978 Free PMC article.
-
Detection of signature sequences in overlapping genes and prediction of a novel overlapping gene in hepatitis G virus.J Mol Evol. 2000 Mar;50(3):284-95. doi: 10.1007/s002399910033. J Mol Evol. 2000. PMID: 10754072 Free PMC article.
-
Molecular evolution in papova viruses and in bacteriophages.Adv Biophys. 1982;15:1-17. doi: 10.1016/0065-227x(82)90003-x. Adv Biophys. 1982. PMID: 6285681 Review.
Cited by
-
The HIV-1 Antisense Gene ASP: The New Kid on the Block.Vaccines (Basel). 2021 May 17;9(5):513. doi: 10.3390/vaccines9050513. Vaccines (Basel). 2021. PMID: 34067514 Free PMC article. Review.
-
New insights into the evolutionary features of viral overlapping genes by discriminant analysis.Virology. 2020 Jul;546:51-66. doi: 10.1016/j.virol.2020.03.007. Epub 2020 Apr 2. Virology. 2020. PMID: 32452417 Free PMC article.
-
Identification of positive and negative regulators of antiviral RNA interference in Arabidopsis thaliana.Nat Commun. 2022 May 30;13(1):2994. doi: 10.1038/s41467-022-30771-0. Nat Commun. 2022. PMID: 35637208 Free PMC article.
-
Coronavirus, the King Who Wanted More Than a Crown: From Common to the Highly Pathogenic SARS-CoV-2, Is the Key in the Accessory Genes?Front Microbiol. 2021 Jul 14;12:682603. doi: 10.3389/fmicb.2021.682603. eCollection 2021. Front Microbiol. 2021. PMID: 34335504 Free PMC article. Review.
-
Prediction of two novel overlapping ORFs in the genome of SARS-CoV-2.Virology. 2021 Oct;562:149-157. doi: 10.1016/j.virol.2021.07.011. Epub 2021 Jul 28. Virology. 2021. PMID: 34339929 Free PMC article.
References
-
- Atkins J.F., Steitz J.A., Anderson C.W., Model P. Binding of mammalian ribosomes to MS2 phage RNA reveals an overlapping gene encoding a lysis function. Cell. 1979;18:247–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

