Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation
- PMID: 31004987
- PMCID: PMC7125799
- DOI: 10.1016/j.virol.2019.03.017
Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation
Abstract
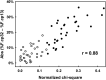
Overlapping genes represent an intriguing puzzle, as they encode two proteins whose ability to evolve is constrained by each other. Overlapping genes can undergo "symmetric evolution" (similar selection pressures on the two proteins) or "asymmetric evolution" (significantly different selection pressures on the two proteins). By sequence analysis of 75 pairs of homologous viral overlapping genes, I evaluated their accordance with one or the other model. Analysis of nucleotide and amino acid sequences revealed that half of overlaps undergo asymmetric evolution, as the protein from one frame shows a number of substitutions significantly higher than that of the protein from the other frame. Interestingly, the most variable protein (often known to interact with the host proteins) appeared to be encoded by the de novo frame in all cases examined. These findings suggest that overlapping genes, besides to increase the coding ability of viruses, are also a source of selective protein adaptation.
Keywords: Ancestral frame; De novo frame; Homologs; Non-synonymous nucleotide substitution; Selection pressure; Synonymous nucleotide substitution; Virus adaptation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
None.
Figures
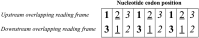
References
-
- Atkins J.F., Steitz J.A., Anderson C.W., Model P. Binding of mammalian ribosomes to MS2 phage RNA reveals an overlapping gene encoding a lysis function. Cell. 1979;18:247–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

