Chronic heavy drinking drives distinct transcriptional and epigenetic changes in splenic macrophages
- PMID: 31005514
- PMCID: PMC6557917
- DOI: 10.1016/j.ebiom.2019.04.027
Chronic heavy drinking drives distinct transcriptional and epigenetic changes in splenic macrophages
Abstract
Background: Chronic heavy alcohol drinking (CHD) leads to significant organ damage, increased susceptibility to infections, and delayed wound healing. These adverse outcomes are believed to be mediated by alterations in the function of myeloid cells; however, the mechanisms underlying these changes are poorly understood.
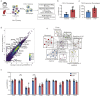
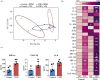
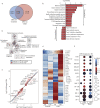
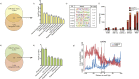
Methods: We determined the impact of CHD on the phenotype of splenic macrophages using flow cytometry. Changes in functional responses to LPS were measured using luminex and RNA-Seq. Finally, alterations in chromatin accessibility were uncovered using ATAC-Seq.
Findings: A history of CHD led to increased frequency of splenic macrophages that exhibited a heightened activation state at resting. Additionally, splenic macrophages from CHD animals generated a larger inflammatory response to LPS, both at protein and gene expression levels. Finally, CHD resulted in increased levels of H3K4me3, a histone mark of active promoters, as well as chromatin accessibility at promoters and intergenic regions that regulate inflammatory responses.
Interpretation: These findings suggest that a history of CHD alters the immune fitness of tissue-resident macrophages via epigenetic mechanisms. FUND: National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institutes of Health (NIH) - R24AA019431, U01 AA13641, U01 AA13510, R21AA021947, and R21AA025839.
Keywords: Chromatin accessibility; Chronic heavy drinking; Ethanol; LPS; Rhesus macaques; Splenic macrophages.
Copyright © 2019. Published by Elsevier B.V.
Figures

Similar articles
-
Transcriptional, Epigenetic, and Functional Reprogramming of Monocytes From Non-Human Primates Following Chronic Alcohol Drinking.Front Immunol. 2021 Aug 20;12:724015. doi: 10.3389/fimmu.2021.724015. eCollection 2021. Front Immunol. 2021. PMID: 34489976 Free PMC article.
-
Dose-dependent effects of chronic alcohol drinking on peripheral immune responses.Sci Rep. 2019 May 24;9(1):7847. doi: 10.1038/s41598-019-44302-3. Sci Rep. 2019. PMID: 31127176 Free PMC article.
-
Ethanol Consumption Induces Nonspecific Inflammation and Functional Defects in Alveolar Macrophages.Am J Respir Cell Mol Biol. 2022 Jul;67(1):112-124. doi: 10.1165/rcmb.2021-0346OC. Am J Respir Cell Mol Biol. 2022. PMID: 35380939 Free PMC article.
-
Transcriptional and Epigenetic Regulation of Monocyte and Macrophage Dysfunction by Chronic Alcohol Consumption.Front Immunol. 2022 Jun 29;13:911951. doi: 10.3389/fimmu.2022.911951. eCollection 2022. Front Immunol. 2022. PMID: 35844518 Free PMC article. Review.
-
Epigenetic basis of the dark side of alcohol addiction.Neuropharmacology. 2017 Aug 1;122:74-84. doi: 10.1016/j.neuropharm.2017.02.002. Epub 2017 Feb 4. Neuropharmacology. 2017. PMID: 28174112 Free PMC article. Review.
Cited by
-
Epigenetics of alcohol-related liver diseases.JHEP Rep. 2022 Mar 10;4(5):100466. doi: 10.1016/j.jhepr.2022.100466. eCollection 2022 May. JHEP Rep. 2022. PMID: 35462859 Free PMC article. Review.
-
Integrated single cell analysis shows chronic alcohol drinking disrupts monocyte differentiation in the bone marrow.Stem Cell Reports. 2023 Sep 12;18(9):1884-1897. doi: 10.1016/j.stemcr.2023.08.001. Epub 2023 Aug 31. Stem Cell Reports. 2023. PMID: 37657446 Free PMC article.
-
Chronic alcohol consumption enhances the differentiation capacity of hematopoietic stem and progenitor cells into osteoclast precursors.bioRxiv [Preprint]. 2025 Feb 8:2025.02.05.636743. doi: 10.1101/2025.02.05.636743. bioRxiv. 2025. Update in: Am J Pathol. 2025 Jul 17:S0002-9440(25)00244-5. doi: 10.1016/j.ajpath.2025.06.010. PMID: 39975302 Free PMC article. Updated. Preprint.
-
Summary of the 2019 alcohol and immunology research interest group (AIRIG) meeting: Alcohol-mediated mechanisms of multiple organ injury.Alcohol. 2020 Sep;87:89-95. doi: 10.1016/j.alcohol.2020.04.008. Epub 2020 Apr 28. Alcohol. 2020. PMID: 32353591 Free PMC article. Review.
-
Integrated single cell analysis shows chronic alcohol drinking disrupts monocyte differentiation in the bone marrow niche.bioRxiv [Preprint]. 2023 Mar 30:2023.03.29.534727. doi: 10.1101/2023.03.29.534727. bioRxiv. 2023. Update in: Stem Cell Reports. 2023 Sep 12;18(9):1884-1897. doi: 10.1016/j.stemcr.2023.08.001. PMID: 37034734 Free PMC article. Updated. Preprint.
References
-
- Schmidt W., De Lint J. Causes of death of alcoholics. Q J Stud Alcohol. 1972;33(1):171–185. - PubMed
-
- Saitz R., Ghali W.A., Moskowitz M.A. The impact of alcohol-related diagnoses on pneumonia outcomes. Arch Intern Med. 1997;157(13):1446–1452. - PubMed
-
- Sabot G., Vendrame G. Incidence of pulmonary tuberculosis in alcoholics. Study based on investigations made at the Ospedale Psichiatrico Provinciale di Udine in the decade 1958-1967. Minerva Med. 1969;60(101):5190–5194. - PubMed
-
- Hudolin V. Tuberculosis and alcoholism. Ann N Y Acad Sci. 1975;252:353–364. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

