A Transient Pulse of Genetic Admixture from the Crusaders in the Near East Identified from Ancient Genome Sequences
- PMID: 31006515
- PMCID: PMC6506814
- DOI: 10.1016/j.ajhg.2019.03.015
A Transient Pulse of Genetic Admixture from the Crusaders in the Near East Identified from Ancient Genome Sequences
Abstract
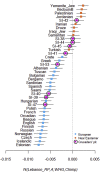
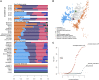
During the medieval period, hundreds of thousands of Europeans migrated to the Near East to take part in the Crusades, and many of them settled in the newly established Christian states along the Eastern Mediterranean coast. Here, we present a genetic snapshot of these events and their aftermath by sequencing the whole genomes of 13 individuals who lived in what is today known as Lebanon between the 3rd and 13th centuries CE. These include nine individuals from the "Crusaders' pit" in Sidon, a mass burial in South Lebanon identified from the archaeology as the grave of Crusaders killed during a battle in the 13th century CE. We show that all of the Crusaders' pit individuals were males; some were Western Europeans from diverse origins, some were locals (genetically indistinguishable from present-day Lebanese), and two individuals were a mixture of European and Near Eastern ancestries, providing direct evidence that the Crusaders admixed with the local population. However, these mixtures appear to have had limited genetic consequences since signals of admixture with Europeans are not significant in any Lebanese group today-in particular, Lebanese Christians are today genetically similar to local people who lived during the Roman period which preceded the Crusades by more than four centuries.
Keywords: Lebanon; Roman period; Sidon; aDNA; medieval period; population history; whole-genome sequences.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Maalouf A. Al Saqi; London: 1984. The Crusades through Arab Eyes.
-
- MacEvitt C.H. University of Pennsylvania Press; 2008. The Crusades and the Christian World of the East: Rough Tolerance.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

