Aurora B-INCENP Localization at Centromeres/Inner Kinetochores Is Required for Chromosome Bi-orientation in Budding Yeast
- PMID: 31006569
- PMCID: PMC6509284
- DOI: 10.1016/j.cub.2019.03.051
Aurora B-INCENP Localization at Centromeres/Inner Kinetochores Is Required for Chromosome Bi-orientation in Budding Yeast
Abstract
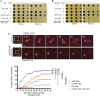
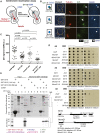
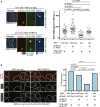
For proper chromosome segregation in mitosis, sister kinetochores must interact with microtubules from opposite spindle poles (chromosome bi-orientation) [1, 2]. To promote bi-orientation, Aurora B kinase disrupts aberrant kinetochore-microtubule interactions [3-6]. It has long been debated how Aurora B halts this action when bi-orientation is established and tension is applied across sister kinetochores. A popular explanation for it is that, upon bi-orientation, sister kinetochores are pulled in opposite directions, stretching the outer kinetochores [7, 8] and moving Aurora B substrates away from Aurora-B-localizing sites at centromeres (spatial separation model) [3, 5, 9]. This model predicts that Aurora B localization at centromeres is required for bi-orientation. However, this notion was challenged by the observation that Bir1 (yeast survivin), which recruits Ipl1-Sli15 (yeast Aurora B-INCENP) to centromeres, can become dispensable for bi-orientation [10]. This raised the possibility that Aurora B localization at centromeres is dispensable for bi-orientation. Alternatively, there might be a Bir1-independent mechanism for recruiting Ipl1-Sli15 to centromeres or inner kinetochores [5, 9]. Here, we show that the COMA inner kinetochore sub-complex physically interacts with Sli15, recruits Ipl1-Sli15 to the inner kinetochore, and promotes chromosome bi-orientation, independently of Bir1, in budding yeast. Moreover, using an engineered recruitment of Ipl1-Sli15 to the inner kinetochore when both Bir1 and COMA are defective, we show that localization of Ipl1-Sli15 at centromeres or inner kinetochores is required for bi-orientation. Our results give important insight into how Aurora B disrupts kinetochore-microtubule interaction in a tension-dependent manner to promote chromosome bi-orientation.
Keywords: Aurora B; Bir1; COMA; INCENP; Ipl1; Mcm21; Sli15; chromosome bi-orientation; kinetochore; survivin.
Copyright © 2019 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures

References
-
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

