MORC3 Is a Target of the Influenza A Viral Protein NS1
- PMID: 31006586
- PMCID: PMC11514443
- DOI: 10.1016/j.str.2019.03.015
MORC3 Is a Target of the Influenza A Viral Protein NS1
Abstract
Microrchidia 3 (MORC3), a human ATPase linked to several autoimmune disorders, has been characterized both as a negative and positive regulator of influenza A virus. Here, we report that the CW domain of MORC3 (MORC3-CW) is targeted by the C-terminal tail of the influenza H3N2 protein NS1. The crystal structure of the MORC3-CW:NS1 complex shows that NS1 occupies the same binding site in CW that is normally occupied by histone H3, a physiological ligand of MORC3-CW. Comparable binding affinities of MORC3-CW to H3 and NS1 peptides and to the adjacent catalytic ATPase domain suggest that the viral protein can compete with the host histone for the association with CW, releasing MORC3 autoinhibition and activating the catalytic function of MORC3. Our structural, biochemical, and cellular analyses suggest that MORC3 might affect the infectivity of influenza virus and therefore has a role in cell immune response.
Keywords: ATPase; CW; MORC3; NS1; chromatin; histone; immune; reader; structure; virus.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


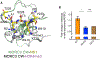
Comment in
-
A Viral Protein Mimics Histone to Hijack Host MORC3.Structure. 2019 Jun 4;27(6):883-885. doi: 10.1016/j.str.2019.05.007. Structure. 2019. PMID: 31167123
Similar articles
-
Mechanism for autoinhibition and activation of the MORC3 ATPase.Proc Natl Acad Sci U S A. 2019 Mar 26;116(13):6111-6119. doi: 10.1073/pnas.1819524116. Epub 2019 Mar 8. Proc Natl Acad Sci U S A. 2019. PMID: 30850548 Free PMC article.
-
Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin.Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):E5108-16. doi: 10.1073/pnas.1609709113. Epub 2016 Aug 15. Proc Natl Acad Sci U S A. 2016. PMID: 27528681 Free PMC article.
-
Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase.Cell Rep. 2016 Sep 20;16(12):3195-3207. doi: 10.1016/j.celrep.2016.08.050. Cell Rep. 2016. PMID: 27653685 Free PMC article.
-
The influenza virus NS1 protein as a therapeutic target.Antiviral Res. 2013 Sep;99(3):409-16. doi: 10.1016/j.antiviral.2013.06.005. Epub 2013 Jun 21. Antiviral Res. 2013. PMID: 23796981 Free PMC article. Review.
-
[Study progress of NS1 protein of influenza A virus].Wei Sheng Wu Xue Bao. 2007 Aug;47(4):729-33. Wei Sheng Wu Xue Bao. 2007. PMID: 17944383 Review. Chinese.
Cited by
-
Non-histone binding functions of PHD fingers.Trends Biochem Sci. 2023 Jul;48(7):610-617. doi: 10.1016/j.tibs.2023.03.005. Epub 2023 Apr 14. Trends Biochem Sci. 2023. PMID: 37061424 Free PMC article. Review.
-
Mechanistic similarities in recognition of histone tails and DNA by epigenetic readers.Curr Opin Struct Biol. 2021 Dec;71:1-6. doi: 10.1016/j.sbi.2021.04.003. Epub 2021 May 13. Curr Opin Struct Biol. 2021. PMID: 33993059 Free PMC article. Review.
-
Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions.Biochem J. 2021 May 28;478(10):1943-1958. doi: 10.1042/BCJ20210203. Biochem J. 2021. PMID: 33969871 Free PMC article.
-
Antiviral effects of interferon-stimulated genes in bats.Front Cell Infect Microbiol. 2023 Aug 18;13:1224532. doi: 10.3389/fcimb.2023.1224532. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37661999 Free PMC article. Review.
-
Structural Investigations of Interactions between the Influenza a Virus NS1 and Host Cellular Proteins.Viruses. 2023 Oct 7;15(10):2063. doi: 10.3390/v15102063. Viruses. 2023. PMID: 37896840 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

