A comprehensive reference transcriptome resource for the Iberian ribbed newt Pleurodeles waltl, an emerging model for developmental and regeneration biology
- PMID: 31006799
- PMCID: PMC6589553
- DOI: 10.1093/dnares/dsz003
A comprehensive reference transcriptome resource for the Iberian ribbed newt Pleurodeles waltl, an emerging model for developmental and regeneration biology
Abstract
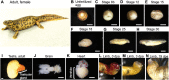
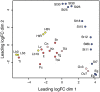
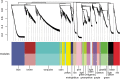
Urodele newts have unique biological properties, notably including prominent regeneration ability. The Iberian ribbed newt, Pleurodeles waltl, is a promising model amphibian distinguished by ease of breeding and efficient transgenic and genome editing methods. However, limited genetic information is available for P. waltl. We conducted an intensive transcriptome analysis of P. waltl using RNA-sequencing to build and annotate gene models. We generated 1.2 billion Illumina reads from a wide variety of samples across 12 different tissues/organs, unfertilized egg, and embryos at eight different developmental stages. These reads were assembled into 1,395,387 contigs, from which 202,788 non-redundant ORF models were constructed. The set is expected to cover a large fraction of P. waltl protein-coding genes, as confirmed by BUSCO analysis, where 98% of universal single-copy orthologs were identified. Ortholog analyses revealed the gene repertoire evolution of urodele amphibians. Using the gene set as a reference, gene network analysis identified regeneration-, developmental-stage-, and tissue-specific co-expressed gene modules. Our transcriptome resource is expected to enhance future research employing this emerging model animal for regeneration research as well as for investigations in other areas including developmental biology, stem cell biology, and cancer research. These data are available via our portal website, iNewt (http://www.nibb.ac.jp/imori/main/).
Keywords: Iberian ribbed newt; NGS; model organism; transcriptome.
© The Author(s) 2019. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures

Similar articles
-
Gene manipulation for regenerative studies using the Iberian ribbed newt, Pleurodeles waltl.Methods Mol Biol. 2015;1290:297-305. doi: 10.1007/978-1-4939-2495-0_23. Methods Mol Biol. 2015. PMID: 25740495
-
Photoperiod-independent testicular development in the model newt Pleurodeles waltl.Dev Growth Differ. 2021 Aug;63(6):277-284. doi: 10.1111/dgd.12738. Epub 2021 Jul 7. Dev Growth Differ. 2021. PMID: 34133763
-
Molecular genetic system for regenerative studies using newts.Dev Growth Differ. 2013 Feb;55(2):229-36. doi: 10.1111/dgd.12019. Epub 2013 Jan 11. Dev Growth Differ. 2013. PMID: 23305125
-
Insights regarding skin regeneration in non-amniote vertebrates: Skin regeneration without scar formation and potential step-up to a higher level of regeneration.Semin Cell Dev Biol. 2020 Apr;100:109-121. doi: 10.1016/j.semcdb.2019.11.014. Epub 2019 Dec 9. Semin Cell Dev Biol. 2020. PMID: 31831357 Review.
-
Contribution of the urodele amphibian Pleurodeles waltl to the analysis of spaceflight-associated immune system deregulation.Mol Immunol. 2013 Dec;56(4):434-41. doi: 10.1016/j.molimm.2013.06.011. Epub 2013 Aug 1. Mol Immunol. 2013. PMID: 23911399 Review.
Cited by
-
FGF signalling plays similar roles in development and regeneration of the skeleton in the brittle star Amphiura filiformis.Development. 2021 May 15;148(10):dev180760. doi: 10.1242/dev.180760. Epub 2021 May 27. Development. 2021. PMID: 34042967 Free PMC article.
-
Comparison of hypoxia- and hyperoxia-induced alteration of epigene expression pattern in lungs of Pleurodeles waltl and Mus musculus.PLoS One. 2024 Feb 28;19(2):e0299661. doi: 10.1371/journal.pone.0299661. eCollection 2024. PLoS One. 2024. PMID: 38416753 Free PMC article.
-
The Axolotl's journey to the modern molecular era.Curr Top Dev Biol. 2022;147:631-658. doi: 10.1016/bs.ctdb.2021.12.010. Epub 2022 Mar 15. Curr Top Dev Biol. 2022. PMID: 35337465 Free PMC article.
-
Newt regeneration genes regulate Wingless signaling to restore patterning in Drosophila eye.iScience. 2021 Sep 24;24(10):103166. doi: 10.1016/j.isci.2021.103166. eCollection 2021 Oct 22. iScience. 2021. PMID: 34746690 Free PMC article.
-
Towards comparative analyses of salamander limb regeneration.J Exp Zool B Mol Dev Evol. 2021 Mar;336(2):129-144. doi: 10.1002/jez.b.22902. Epub 2019 Oct 4. J Exp Zool B Mol Dev Evol. 2021. PMID: 31584252 Free PMC article. Review.
References
-
- Spemann H., Mangold O.. 1924, Über Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren, Archiv Mikr. Anat. u Entwicklungsmechanik, 100, 599–638.
-
- Wolff G. 1895, Entwicklungsphysiologische Studien 1. Die regeneration der Urodelenlinse, Wilhelm Roux’s Arch, Entwined Meghan Org., 1, 380–90.
-
- Agata K., Inoue T.. 2012, Survey of the differences between regenerative and non-regenerative animals, Dev. Growth Differ., 54, 143–52. - PubMed
-
- Hayashi T., Mizuno N., Kondoh H.. 2008, Determinative roles of FGF and Wnt signals in iris-derived lens regeneration in newt eye, Dev. Growth Differ., 50, 279–87. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

