Maturation of atypical ribosomal RNA precursors in Helicobacter pylori
- PMID: 31006803
- PMCID: PMC6582327
- DOI: 10.1093/nar/gkz258
Maturation of atypical ribosomal RNA precursors in Helicobacter pylori
Abstract
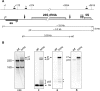
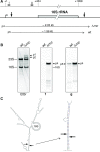
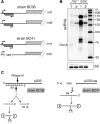
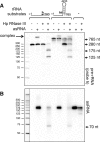
In most bacteria, ribosomal RNA is transcribed as a single polycistronic precursor that is first processed by RNase III. This double-stranded specific RNase cleaves two large stems flanking the 23S and 16S rRNA mature sequences, liberating three 16S, 23S and 5S rRNA precursors, which are further processed by other ribonucleases. Here, we investigate the rRNA maturation pathway of the human gastric pathogen Helicobacter pylori. This bacterium has an unusual arrangement of its rRNA genes, the 16S rRNA gene being separated from a 23S-5S rRNA cluster. We show that RNase III also initiates processing in this organism, by cleaving two typical stem structures encompassing 16S and 23S rRNAs and an atypical stem-loop located upstream of the 5S rRNA. Deletion of RNase III leads to the accumulation of a large 23S-5S precursor that is found in polysomes, suggesting that it can function in translation. Finally, we characterize a cis-encoded antisense RNA overlapping the leader of the 23S-5S rRNA precursor. We present evidence that this antisense RNA interacts with this precursor, forming an intermolecular complex that is cleaved by RNase III. This pairing induces additional specific cleavages of the rRNA precursor coupled with a rapid degradation of the antisense RNA.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis.Mol Microbiol. 2008 Jun;68(5):1096-106. doi: 10.1111/j.1365-2958.2008.06207.x. Epub 2008 Mar 19. Mol Microbiol. 2008. PMID: 18363798
-
RNase III Processing of rRNA in the Lyme Disease Spirochete Borrelia burgdorferi.J Bacteriol. 2018 Jun 11;200(13):e00035-18. doi: 10.1128/JB.00035-18. Print 2018 Jul 1. J Bacteriol. 2018. PMID: 29632096 Free PMC article.
-
Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T.RNA. 1999 Jan;5(1):139-46. doi: 10.1017/s1355838299981669. RNA. 1999. PMID: 9917073 Free PMC article.
-
Intermolecular base-paired interaction between complementary sequences present near the 3' ends of 5S rRNA and 18S (16S) rRNA might be involved in the reversible association of ribosomal subunits.Nucleic Acids Res. 1979 Dec 11;7(7):1913-29. doi: 10.1093/nar/7.7.1913. Nucleic Acids Res. 1979. PMID: 94160 Free PMC article. Review.
-
Mechanism and regulation of bacterial ribosomal RNA processing.Annu Rev Microbiol. 1990;44:105-29. doi: 10.1146/annurev.mi.44.100190.000541. Annu Rev Microbiol. 1990. PMID: 1701293 Review. No abstract available.
Cited by
-
Enhanced Symbiotic Characteristics in Bacterial Genomes with the Disruption of rRNA Operon.Biology (Basel). 2020 Dec 3;9(12):440. doi: 10.3390/biology9120440. Biology (Basel). 2020. PMID: 33287185 Free PMC article.
-
Specialised ribosomes as versatile regulators of gene expression.RNA Biol. 2022 Jan;19(1):1103-1114. doi: 10.1080/15476286.2022.2135299. RNA Biol. 2022. PMID: 36255182 Free PMC article. Review.
-
Unlinked rRNA genes are widespread among bacteria and archaea.ISME J. 2020 Feb;14(2):597-608. doi: 10.1038/s41396-019-0552-3. Epub 2019 Nov 11. ISME J. 2020. PMID: 31712737 Free PMC article.
-
RNase III, Ribosome Biogenesis and Beyond.Microorganisms. 2021 Dec 17;9(12):2608. doi: 10.3390/microorganisms9122608. Microorganisms. 2021. PMID: 34946208 Free PMC article. Review.
-
Study of key RNA metabolism proteins in Enterococcus faecalis.RNA Biol. 2020 Jun;17(6):794-804. doi: 10.1080/15476286.2020.1728103. Epub 2020 Feb 19. RNA Biol. 2020. PMID: 32070211 Free PMC article.
References
-
- Srivastava A.K., Schlessinger D.. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 1990; 44:105–129. - PubMed
-
- Bram R.J., Young R.A., Steitz J.A.. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980; 19:393–401. - PubMed
-
- Herskovitz M.A., Bechhofer D.H.. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol. 2000; 38:1027–1033. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

