Perturbation of mRNP biogenesis reveals a dynamic landscape of the Rrp6-dependent surveillance machinery trafficking along the yeast genome
- PMID: 31007122
- PMCID: PMC6546349
- DOI: 10.1080/15476286.2019.1593745
Perturbation of mRNP biogenesis reveals a dynamic landscape of the Rrp6-dependent surveillance machinery trafficking along the yeast genome
Abstract
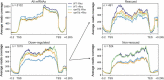
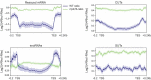
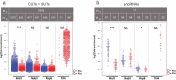
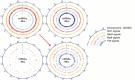
Eukaryotic cells have evolved a nuclear quality control (QC) system to monitor the co-transcriptional mRNA processing and packaging reactions that lead to the formation of export-competent ribonucleoprotein particles (mRNPs). Aberrant mRNPs that fail to pass the QC steps are retained in the nucleus and eliminated by the exonuclease activity of Rrp6. It is still unclear how the surveillance system is precisely coordinated both physically and functionally with the transcription machinery to detect the faulty events that may arise at each step of transcript elongation and mRNP formation. To dissect the QC mechanism, we previously implemented a powerful assay based on global perturbation of mRNP biogenesis in yeast by the bacterial Rho helicase. By monitoring model genes, we have shown that the QC process is coordinated by Nrd1, a component of the NNS complex (Nrd1-Nab3-Sen1) involved in termination, processing and decay of ncRNAs which is recruited by the CTD of RNAP II. Here, we have extended our investigations by analyzing the QC behaviour over the whole yeast genome. We performed high-throughput RNA sequencing (RNA-seq) to survey a large collection of mRNPs whose biogenesis is affected by Rho action and which can be rescued upon Rrp6 depletion. This genome-wide perspective was extended by generating high-resolution binding landscapes (ChIP-seq) of QC components along the yeast chromosomes before and after perturbation of mRNP biogenesis. Our results show that perturbation of mRNP biogenesis redistributes the QC components over the genome with a significant hijacking of Nrd1 and Nab3 from genomic loci producing ncRNAs to Rho-affected protein-coding genes, triggering termination and processing defects of ncRNAs.
Keywords: ChIP-seq; RNA-seq; mRNP biogenesis; mRNP quality control; yeast.
Figures



References
-
- Luna R, Gaillard H, Gonzalez-Aguilera C, et al. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. - PubMed
-
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. - PubMed
-
- Fasken MB, Corbett AH. Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol. 2005;12:482–488. - PubMed
-
- Schmid M, Jensen TH. Transcription-associated quality control of mRNP. Biochim Biophys Acta. 2013;1829:158–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
