MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint
- PMID: 31007149
- PMCID: PMC6984599
- DOI: 10.1080/15548627.2019.1606647
MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint
Abstract
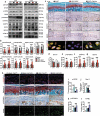
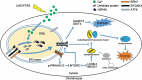
A switch from autophagy to apoptosis is implicated in chondrocytes during the osteoarthritis (OA) progression with currently unknown mechanism(s). In this study we utilized a flow fluid shear stress (FFSS) model in cultured chondrocytes and a unilateral anterior crossbite (UAC) animal model. We found that both FFSS and UAC actively induced endoplasmic reticulum stress (ERS) in the temporomandibular joints (TMJ) chondrocytes, as demonstrated by dramatic increases in expression of HSPA5, p-EIF2AK3, p-ERN1 and ATF6. Interestingly, both FFSS and UAC activated not only pro-death p-EIF2AK3-mediated ERS-apoptosis programs but also pro-survival p-ERN1-mediated autophagic flux in chondrocytes. Data from FFSS demonstrated that MTORC1, a downstream of p-ERN1, suppressed autophagy but promoted p-EIF2AK3 mediated ERS-apoptosis. Data from UAC model demonstrated that at early stage both the p-ERN1 and p-EIF2AK3 were activated and MTORC1 was inhibited in TMJ chondrocytes. At late stage, MTORC1-p-EIF2AK3-mediated ERS apoptosis were predominant, while p-ERN1 and autophagic flux were inhibited. Inhibition of MTORC1 by TMJ local injection of rapamycin in rats or inducible ablation of MTORC1 expression selectively in chondrocytes in mice promoted chondrocyte autophagy and suppressed apoptosis, and reduced TMJ cartilage loss induced by UAC. In contrast, MTORC1 activation by TMJ local administration of MHY1485 or genetic deletion of Tsc1, an upstream MTORC1 suppressor, resulted in opposite effects. Collectively, our results establish that aberrant mechanical loading causes cartilage degeneration by activating, at least in part, the MTORC1 signaling which modulates the autophagy and apoptosis programs in TMJ chondrocytes. Thus, inhibition of MTORC1 provides a novel therapeutic strategy for prevention and treatment of OA.Abbreviations : ACTB: actin beta; ATF6: activating transcription factor 6; BECN1: beclin 1; BFL: bafilomycin A1; CASP12: caspase 12; CASP3: caspase 3; DAPI: 4',6-diamidino-2-phenylindole; DDIT3: DNA-damage inducible transcript 3; EIF2AK3/PERK: eukaryotic translation initiation factor 2 alpha kinase 3; ER: endoplasmic reticulum; ERS: endoplasmic reticulum stress; ERN1/IRE1: endoplasmic reticulum to nucleus signaling 1; FFSS: flow fluid shear stress; HSPA5/GRP78/BiP: heat shock protein 5; LAMP2: lysosome-associated membrane protein 2; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MTOR: mechanistic target of rapamycin kinase; MTORC1: mechanistic target of rapamycin complex 1; OA: osteoarthritis; PRKAA1/2/AMPK1/2: protein kinase, AMP-activated, alpha 1/2 catalytic subunit; RPS6: ribosomal protein S6; Rapa: rapamycin; SQSTM1/p62: sequestosome 1; TEM: transmission electron microscopy; TG: thapsigargin; TMJ: temporomandibular joints; TSC1/2: tuberous sclerosis complex 1/2; UAC: unilateral anterior crossbite; UPR: unfolded protein response; XBP1: x-box binding protein 1.
Keywords: Autophagy; chondrocyte; endoplasmic reticulum stress; endoplasmic reticulum to nucleus signaling 1; eukaryotic translation initiation factor 2 alpha kinase 3; mechanistic target of rapamycin complex 1; osteoarthritis.
Figures




References
-
- Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376–387. PMID: 25748615. - PubMed
-
- Bijlsma JW, Berenbaum F, Lafeber FP.. Osteoarthritis: an update with relevance for clinical practice. Lancet. 2011;377(9783):2115–2126. PMID: 21684382. - PubMed
-
- Scrivani SJ, Keith DA, Kaban LB.. Temporomandibular disorders. N Engl J Med. 2008;359(25):2693–2705. PMID: 19092154. - PubMed
-
- Zhang M, Yang H, Lu L, et al. Matrix replenishing by BMSCs is beneficial for osteoarthritic temporomandiblar joint cartilage. Osteoarthritis Cartilage. 2017;25(9):1551–1562. PMID: 28532603. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
