Oncogenic Role of ZFAS1 lncRNA in Head and Neck Squamous Cell Carcinomas
- PMID: 31010087
- PMCID: PMC6523746
- DOI: 10.3390/cells8040366
Oncogenic Role of ZFAS1 lncRNA in Head and Neck Squamous Cell Carcinomas
Abstract
Background: Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease with high mortality. The identification of specific HNSCC biomarkers will increase treatment efficacy and limit the toxicity of current therapeutic strategies. Long non-coding RNAs (lncRNAs) are promising biomarkers. Accordingly, here we investigate the biological role of ZFAS1 and its potential as a biomarker in HNSCC.
Methods: The expression level of ZFAS1 in HNSCC cell lines was analyzed using qRT-PCR. Based on the HNSCC TCGA data, the ZFAS1 expression profile, clinicopathological features, and expression of correlated genes were analyzed in patient tissue samples. The selected genes were classified according to their biological function using the PANTHER tool. The interaction between lncRNA:miRNA and miRNA:mRNA was tested using available online tools. All statistical analyses were accomplished using GraphPad Prism 5.
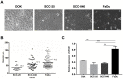
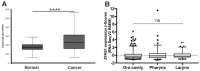
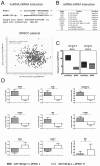
Results: The expression of ZFAS1 was up-regulated in the metastatic FaDu cell line relative to the less aggressive SCC-25 and SCC-040 and dysplastic DOK cell lines. The TCGA data indicated an up-regulation of ZFAS1 in HNSCCs compared to normal tissue samples. The ZFAS1 levels typically differed depending on the cancer stage and T-stage. Patients with a lower expression of ZFAS1 presented a slightly longer disease-free survival and overall survival. The analysis of genes associated with ZFAS1, as well its targets, indicate that they are linked with crucial cellular processes. In the group of patients with low expression of ZFAS1, we detected the up-regulation of suppressors and down-regulation of genes associated with epithelial-to-mesenchymal transition (EMT) process, metastases, and cancer-initiating cells. Moreover, the negative correlation between ZFAS1 and its host gene, ZNFX1, was observed. The analysis of interactions indicated that ZFAS1 has a binding sequence for miR-150-5p. The expression of ZFAS1 and miR-150-5p is negatively correlated in HNSCC patients. miR-150-5p can regulate the 3'UTR of EIF4E mRNA. In the group of patients with high expression of ZFAS1 and low expression of miR-150-5p, we detected an up-regulation of EIF4E.
Conclusions: In HNSCC, ZFAS1 displays oncogenic properties, regulates important processes associated with EMT, cancer-initiating cells, and metastases, and might affect patients' clinical outcomes. ZFAS1 likely regulates the cell phenotype through miR-150-5p and its downstream targets. Following further validation, ZFAS1 might prove a new and valuable biomarker.
Keywords: HNSCC; ZFAS1; ZNFX1 antisense RNA 1; biomarker; head and neck cancers; lncRNA; non-coding RNA.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures
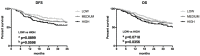

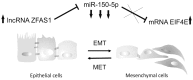
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

