miR-128 Restriction of LINE-1 (L1) Retrotransposition Is Dependent on Targeting hnRNPA1 mRNA
- PMID: 31010097
- PMCID: PMC6515209
- DOI: 10.3390/ijms20081955
miR-128 Restriction of LINE-1 (L1) Retrotransposition Is Dependent on Targeting hnRNPA1 mRNA
Abstract
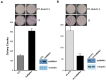
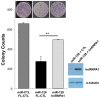
The majority of the human genome is made of transposable elements, giving rise to interspaced repeats, including Long INterspersed Element-1s (LINE-1s or L1s). L1s are active human transposable elements involved in genomic diversity and evolution; however, they can also contribute to genomic instability and diseases. L1s require host factors to complete their life cycles, whereas the host has evolved numerous mechanisms to restrict L1-induced mutagenesis. Restriction mechanisms in somatic cells include methylation of the L1 promoter, anti-viral factors and RNA-mediated processes such as small RNAs. microRNAs (miRNAs or miRs) are small non-coding RNAs that post-transcriptionally repress multiple target genes often found in the same cellular pathways. We have recently established that miR-128 functions as a novel restriction factor inhibiting L1 mobilization in somatic cells. We have further demonstrated that miR-128 functions through a dual mechanism; by directly targeting L1 RNA for degradation and indirectly by inhibiting a cellular co-factor which L1 is dependent on to transpose to new genomic locations (TNPO1). Here, we add another piece to the puzzle of the enigmatic L1 lifecycle. We show that miR-128 also inhibits another key cellular factor, hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1), by significantly reducing mRNA and protein levels through direct interaction with the coding sequence (CDS) of hnRNPA1 mRNA. In addition, we demonstrate that repression of hnRNPA1 using hnRNPA1-shRNA significantly decreases de novo L1 retro-transposition and that induced hnRNPA1 expression enhances L1 mobilization. Furthermore, we establish that hnRNPA1 is a functional target of miR-128. Finally, we determine that induced hnRNPA1 expression in miR-128-overexpressing cells can partly rescue the miR-128-induced repression of L1's ability to transpose to different genomic locations. Thus, we have identified an additional mechanism by which miR-128 represses L1 retro-transposition and mediates genomic stability.
Keywords: LINE-1; hnRNPA1; miR-128; miRs; restriction factor; retrotransposition.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
MicroRNA miR-128 represses LINE-1 (L1) retrotransposition by down-regulating the nuclear import factor TNPO1.J Biol Chem. 2017 Dec 15;292(50):20494-20508. doi: 10.1074/jbc.M117.807677. Epub 2017 Oct 3. J Biol Chem. 2017. PMID: 28974576 Free PMC article.
-
miR-128 represses L1 retrotransposition by binding directly to L1 RNA.Nat Struct Mol Biol. 2015 Oct;22(10):824-31. doi: 10.1038/nsmb.3090. Epub 2015 Sep 14. Nat Struct Mol Biol. 2015. PMID: 26367248
-
LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression.RNA Biol. 2020 Mar;17(3):381-394. doi: 10.1080/15476286.2019.1708547. Epub 2020 Jan 2. RNA Biol. 2020. PMID: 31868085 Free PMC article.
-
From the genome's perspective: Bearing somatic retrotransposition to leverage the regulatory potential of L1 RNAs.Bioessays. 2025 Feb;47(2):e2400125. doi: 10.1002/bies.202400125. Epub 2024 Nov 9. Bioessays. 2025. PMID: 39520370 Free PMC article. Review.
-
The multifarious roles of heterogeneous ribonucleoprotein A1 in viral infections.Rev Med Virol. 2020 Mar;30(2):e2097. doi: 10.1002/rmv.2097. Epub 2020 Jan 27. Rev Med Virol. 2020. PMID: 31989716 Free PMC article. Review.
Cited by
-
Upregulation of the heterogeneous nuclear ribonucleoprotein hnRNPA1 is an independent predictor of early biochemical recurrence in TMPRSS2:ERG fusion-negative prostate cancers.Virchows Arch. 2020 Nov;477(5):625-636. doi: 10.1007/s00428-020-02834-4. Epub 2020 May 16. Virchows Arch. 2020. PMID: 32417965 Free PMC article.
-
SLFN11 Restricts LINE-1 Mobility.Cells. 2025 May 28;14(11):790. doi: 10.3390/cells14110790. Cells. 2025. PMID: 40497966 Free PMC article.
-
The Role of Transposable Elements of the Human Genome in Neuronal Function and Pathology.Int J Mol Sci. 2022 May 23;23(10):5847. doi: 10.3390/ijms23105847. Int J Mol Sci. 2022. PMID: 35628657 Free PMC article. Review.
-
The oncogenic microRNA miR-222 promotes human LINE-1 retrotransposition.RNA Biol. 2025 Dec;22(1):1-15. doi: 10.1080/15476286.2025.2511318. Epub 2025 Jun 3. RNA Biol. 2025. PMID: 40421600 Free PMC article.
-
Recognize Yourself-Innate Sensing of Non-LTR Retrotransposons.Viruses. 2021 Jan 12;13(1):94. doi: 10.3390/v13010094. Viruses. 2021. PMID: 33445593 Free PMC article. Review.
References
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

