Studies Towards Hypoxia-Activated Prodrugs of PARP Inhibitors
- PMID: 31010230
- PMCID: PMC6514732
- DOI: 10.3390/molecules24081559
Studies Towards Hypoxia-Activated Prodrugs of PARP Inhibitors
Abstract
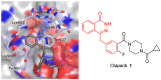
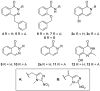
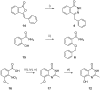
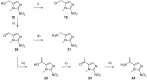
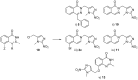
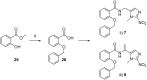
Poly(ADP-ribose)polymerase (PARP) inhibitors (PARPi) have recently been approved for the treatment of breast and ovarian tumors with defects in homologous recombination repair (HRR). Although it has been demonstrated that PARPi also sensitize HRR competent tumors to cytotoxic chemotherapies or radiotherapy, normal cell toxicity has remained an obstacle to their use in this context. Hypoxia-activated prodrugs (HAPs) provide a means to limit exposure of normal cells to active drug, thus adding a layer of tumor selectivity. We have investigated potential HAPs of model PARPi in which we attach a bioreducible "trigger" to the amide nitrogen, thereby blocking key binding interactions. A representative example showed promise in abrogating PARPi enzymatic activity in a biochemical assay, with a ca. 160-fold higher potency of benzyl phthalazinone 4 than the corresponding model HAP 5, but these N-alkylated compounds did not release the PARPi upon one-electron reduction by radiolysis. Therefore, we extended our investigation to include NU1025, a PARPi that contains a phenol distal to the core binding motif. The resulting 2-nitroimidazolyl ether provided modest abrogation of PARPi activity with a ca. seven-fold decrease in potency, but released the PARPi efficiently upon reduction. This investigation of potential prodrug approaches for PARPi has identified a useful prodrug strategy for future exploration.
Keywords: NU1025; PARP inhibitor; anti-cancer agents; hypoxia; hypoxia-activated prodrugs; prodrug; radiolytic reduction; tumor targeting.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; and in the decision to publish the results.
Figures
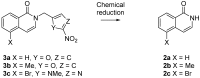


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

