Ancestry patterns inferred from massive RNA-seq data
- PMID: 31010885
- PMCID: PMC6573782
- DOI: 10.1261/rna.070052.118
Ancestry patterns inferred from massive RNA-seq data
Abstract
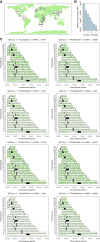
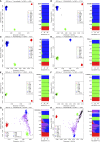
There is a growing body of evidence suggesting that patterns of gene expression vary within and between human populations. However, the impact of this variation in human diseases has been poorly explored, in part owing to the lack of a standardized protocol to estimate biogeographical ancestry from gene expression studies. Here we examine several studies that provide new solid evidence indicating that the ancestral background of individuals impacts gene expression patterns. Next, we test a procedure to infer genetic ancestry from RNA-seq data in 25 data sets where information on ethnicity was reported. Genome data of reference continental populations retrieved from The 1000 Genomes Project were used for comparisons. Remarkably, only eight out of 25 data sets passed FastQC default filters. We demonstrate that, for these eight population sets, the ancestral background of donors could be inferred very efficiently, even in data sets including samples with complex patterns of admixture (e.g., American-admixed populations). For most of the gene expression data sets of suboptimal quality, ancestral inference yielded odd patterns. The present study thus brings a cautionary note for gene expression studies highlighting the importance to control for the potential confounding effect of ancestral genetic background.
Keywords: RNA-seq; SNPs; biogeographical ancestry; gene expression; genomics; transcriptomics.
© 2019 Barral-Arca et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Aung T, Ozaki M, Lee MC, Schlötzer-Schrehardt U, Thorleifsson G, Mizoguchi T, Igo RP Jr, Haripriya A, Williams SE, Astakhov YS, et al. 2017. Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci. Nat Genet 49: 993–1004. 10.1038/ng.3875 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
