Homozygous noncanonical splice variant in LSM1 in two siblings with multiple congenital anomalies and global developmental delay
- PMID: 31010896
- PMCID: PMC6549555
- DOI: 10.1101/mcs.a004101
Homozygous noncanonical splice variant in LSM1 in two siblings with multiple congenital anomalies and global developmental delay
Abstract
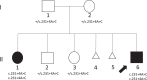
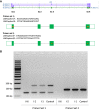
Two siblings, one male and one female, ages 6 and 13 yr old, have similar clinical features of global developmental delay, multiple congenital anomalies affecting the cardiac, genitourinary, and skeletal systems, and abnormal eye movements. Whole-genome sequencing revealed a homozygous splice variant (NM_014462.3:c.231+4A>C) in LSM1 that segregated with the phenotype in the family. LSM1 has a role in pre-mRNA splicing and degradation. Expression studies revealed absence of expression of the canonical isoform in the affected individuals. The Lsm1 knockout mice have a partially overlapping phenotype that affects the brain, heart, and eye. To our knowledge, LSM1 has not been associated with any human disorder; however, the tissue expression pattern, gene constraint, and the similarity of the phenotype in our patients and the knockout mice models suggest it has a role in the development of multiple organ systems in humans.
Keywords: bicuspid aortic valve; bilateral cryptorchidism; congenital mitral stenosis; congenital strabismus; craniofacial asymmetry; cupped ear; hydronephrosis; hydroureter; inguinal hernia; intellectual disability, moderate; intermittent microsaccadic pursuits; lumbar hemivertebrae; moderate global developmental delay; penile hypospadias; perimembranous ventricular septal defect; primum atrial septal defect; triphalangeal thumb.
© 2019 Okur et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
