Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study
- PMID: 31011206
- PMCID: PMC6553618
- DOI: 10.1038/s41591-019-0407-5
Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study
Abstract
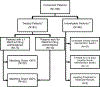
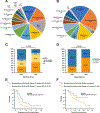
Cancer treatments have evolved from indiscriminate cytotoxic agents to selective genome- and immune-targeted drugs that have transformed the outcomes of some malignancies1. Tumor complexity and heterogeneity suggest that the 'precision medicine' paradigm of cancer therapy requires treatment to be personalized to the individual patient2-6. To date, precision oncology trials have been based on molecular matching with predetermined monotherapies7-14. Several of these trials have been hindered by very low matching rates, often in the 5-10% range15, and low response rates. Low matching rates may be due to the use of limited gene panels, restrictive molecular matching algorithms, lack of drug availability, or the deterioration and death of end-stage patients before therapy can be implemented. We hypothesized that personalized treatment with combination therapies would improve outcomes in patients with refractory malignancies. As a first test of this concept, we implemented a cross-institutional prospective study (I-PREDICT, NCT02534675 ) that used tumor DNA sequencing and timely recommendations for individualized treatment with combination therapies. We found that administration of customized multidrug regimens was feasible, with 49% of consented patients receiving personalized treatment. Targeting of a larger fraction of identified molecular alterations, yielding a higher 'matching score', was correlated with significantly improved disease control rates, as well as longer progression-free and overall survival rates, compared to targeting of fewer somatic alterations. Our findings suggest that the current clinical trial paradigm for precision oncology, which pairs one driver mutation with one drug, may be optimized by treating molecularly complex and heterogeneous cancers with combinations of customized agents.
Conflict of interest statement
COMPETING INTERESTS:
Jason Sicklick receives research funds from Foundation Medicine Inc., Novartis Pharmaceuticals, Blueprint Medicines, and Amgen, as well as consultant fees from Loxo, Biotheranostics, and Grand Rounds. Michael Hahn receives research funds from General Electric and is an equity holder in Illumina, Inc. Casey Williams receives research support from Takeda, Tesaro, and Pfizer, as well as consultant fees from Takeda. Vincent Miller, Jeffrey Ross, and Jennifer Webster are employees and equity holders in Foundation Medicine Inc. Vincent Miller is also on the Board of Directors for Revolution Medicines (equity and compensation >$10,000) and has patent royalties for EGFR T790M testing issued to Memorial Sloan Kettering Cancer Center. Adam Benson is formerly an employee and is an equity holder in Foundation Medicine Inc. He is now an employee of Scipher Medicine. Philip J. Stephens is formerly an employee and is formerly an equity holder in Foundation Medicine Inc. He is now an employee of Grail, Inc. J. Jack Lee served on the Statistical Advisory Board of AbbVie. Razelle Kurzrock has research funding from Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Grifols, Omniseq, Foundation Medicine Inc., Guardant Health, and Konica Minolta, as well as consultant fees from Loxo, Actuate Therapeutics, Roche, Xbiotech and NeoMed. She serves as an advisor to Soluventis. She receives speaker fees from Roche, and has an equity interest in IDbyDNA, Curematch, Inc., and Soluventis. All other authors have no relationships to disclose.
Figures
Comment in
-
Molecular profiling in precision medicine oncology.Nat Med. 2019 May;25(5):711-712. doi: 10.1038/s41591-019-0442-2. Nat Med. 2019. PMID: 31036881 No abstract available.
-
Precision Oncology: Three Small Steps Forward.Cancer Cell. 2019 Jun 10;35(6):825-826. doi: 10.1016/j.ccell.2019.05.009. Cancer Cell. 2019. PMID: 31185208 Free PMC article.
-
Studies Show Clinical Utility of Precision Medicine.Cancer Discov. 2019 Aug;9(8):OF8. doi: 10.1158/2159-8290.CD-ND2019-008. Epub 2019 Jun 26. Cancer Discov. 2019. PMID: 31242995
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

